













| General Information: |
|
| Name(s) found: |
TRS23 /
YDR246W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 219 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TRAPP complex
[IDA |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[IMP |
| Molecular Function: |
Ras guanyl-nucleotide exchange factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIETILVIN KSGGLIYQRN FTNDEQKLNS NEYLILASTL HGVFAIASQL TPKALQLTQQ 60
61 TNIENTIPYI PYVGMSSNRS DTRNGGGNNN KHTNNEKLGS FKGDDFFKEP FTNWNKSGLR 120
121 QLCTDQFTMF IYQTLTGLKF VAISSSVMPQ RQPTIATTDK PDRPKSTSNL AIQIADNFLR 180
181 KVYCLYSDYV MKDPSYSMEM PIRSNLFDEK VKKMVENLQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
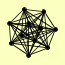
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BET3 BET5 FKS1 GSG1 KRE11 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 |
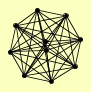
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
BET3 BET5 GSG1 KRE11 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 |
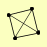
| View Details | Krogan NJ, et al. (2006) |
|
TRS120 TRS23 TRS31 TRS33 |
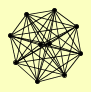
| View Details | Gavin AC, et al. (2006) |
|
BET3 BET5 GSG1 KRE11 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 |
| View Details | Gavin AC, et al. (2002) |
|
BET3 BET5 FKS1 GSG1 KRE11 RVB2 SKN1 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 |
| View Details | Qiu et al. (2008) |
|
BET3 BET5 GSG1 GYP6 KRE11 SED5 TRS120 TRS130 TRS20 TRS23 TRS31 TRS33 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
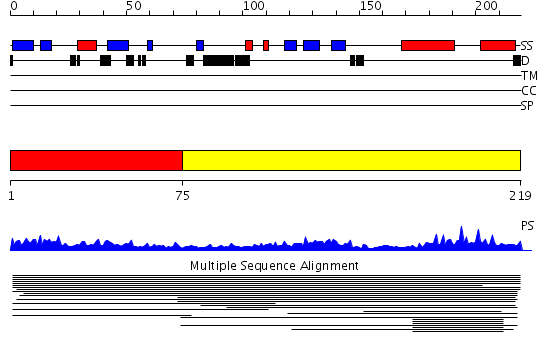
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | 1.015988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [75..219] | 5.016997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)