













| General Information: |
|
| Name(s) found: |
SEC63 /
YOR254C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 663 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[TAS Sec62/Sec63 complex [IPI mitochondrion [IDA |
| Biological Process: |
SRP-dependent cotranslational protein targeting to membrane
[IMP cytosol to ER transport [IMP posttranslational protein targeting to membrane [IMP |
| Molecular Function: |
protein transporter activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTNYEYDEA SETWPSFILT GLLMVVGPMT LLQIYQIFFG ANAEDGNSGK SKEFNEEVFK 60
61 NLNEEYTSDE IKQFRRKFDK NSNKKSKIWS RRNIIIIVGW ILVAILLQRI NSNDAIKDAA 120
121 TKLFDPYEIL GISTSASDRD IKSAYRKLSV KFHPDKLAKG LTPDEKSVME ETYVQITKAY 180
181 ESLTDELVRQ NYLKYGHPDG PQSTSHGIAL PRFLVDGSAS PLLVVCYVAL LGLILPYFVS 240
241 RWWARTQSYT KKGIHNVTAS NFVSNLVNYK PSEIVTTDLI LHWLSFAHEF KQFFPDLQPT 300
301 DFEKLLQDHI NRRDSGKLNN AKFRIVAKCH SLLHGLLDIA CGFRNLDIAL GAINTFKCIV 360
361 QAVPLTPNCQ ILQLPNVDKE HFITKTGDIH TLGKLFTLED AKIGEVLGIK DQAKLNETLR 420
421 VASHIPNLKI IKADFLVPGE NQVTPSSTPY ISLKVLVRSA KQPLIPTSLI PEENLTEPQD 480
481 FESQRDPFAM MSKQPLVPYS FAPFFPTKRR GSWCCLVSSQ KDGKILQTPI IIEKLSYKNL 540
541 NDDKDFFDKR IKMDLTKHEK FDINDWEIGT IKIPLGQPAP ETVGDFFFRV IVKSTDYFTT 600
601 DLDITMNMKV RDSPAVEQVE VYSEEDDEYS TDDDETESDD ESDASDYTDI DTDTEAEDDE 660
661 SPE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SEC62 SEC63 SEC66 SEC72 |
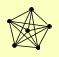
| View Details | Hazbun TR, et al. (2003) |
|
AIM25 DED81 GWT1 MRC1 SEC63 SEC7 |
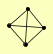
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
SEC62 SEC63 SEC66 SEC72 |
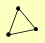
| View Details | Krogan NJ, et al. (2006) |
|
SEC63 SEC66 SEC72 |
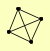
| View Details | Gavin AC, et al. (2006) |
|
SEC62 SEC63 SEC66 SEC72 |
| View Details | Gavin AC, et al. (2002) |
|
ANP1 FKS1 MNN10 SEC62 SEC63 SEC66 SEC72 |
| View Details | Qiu et al. (2008) |
|
SBH1 SBH2 SEC61 SEC62 SEC63 SEC66 SEC72 SSH1 SSS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
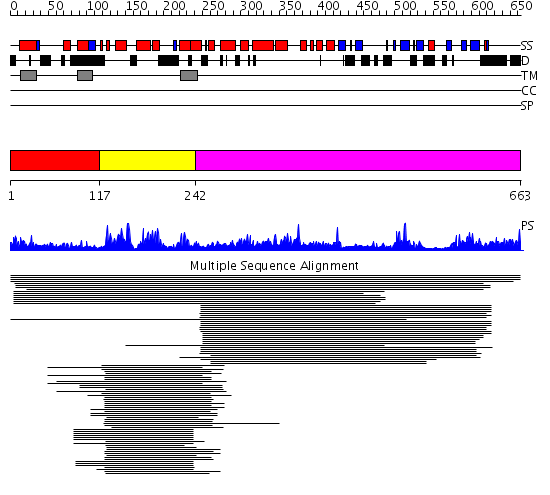
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..241] | 127.502279 | DnaJ chaperone, N-terminal (J) domain | |
| 3 | View Details | [242..663] | 12.139943 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|