













| General Information: |
|
| Name(s) found: |
TCB3 /
YML072C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1545 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud
[IDA mitochondrion [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
lipid binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTGIKAQVHP PPDSTLFHEE EKKKVGGNLP QKVINQQERG SDHAPSGHHQ YHQLINHDAN 60
61 DTKTSNSVSD VSKGQKTADS NPEGKKQSSK DIFVASSAQK TNQLPGPNPQ GSIGAVPLEG 120
121 LRPKEFRSAP SRKPNKFDTS ITKPGVLDDL GKLDEKDIKE KFHLDSDDKL FPWQNVGEFH 180
181 ASGKGSPNTK MSRVIKAYIL ENFYNDWYCN IATVLGTCFF SWLFAYIGFS WWSMIFIFLG 240
241 TATVYNAEYT RFNRNIRDDL KRVTVEETLS DRVESTTWLN SFLSKFWVIY MPVLSQQVKD 300
301 NVNPQLAGVA PGYGIDALAI DEFTLGSKAP TIKGIKSYTK TGKNTVEMDW SFAFTPSDVS 360
361 DMTATEAREK INPKISLGVT LGKSFVSKTM PILVEDINVA GKMRIKVEFG KAFPNIKIVS 420
421 LQLLEPPLID FALKPIGGDT LGLDVMSFLP GLKSFVKNII NSNIGPMLFP PNHLDINVED 480
481 IMAAQSKEAI GVLAVTIASA DSLKGSDFIT NTVDPYIVMT TEDAVPGTDE EVRTSIKSNV 540
541 KNPRWNETKY LLLNTLEQKL NLKCFDFNDV RKDTVIGDLQ LDLADLLQNP VLDNQTAELR 600
601 SGTKSKGILH YSLHWFPVKE DKSEEKAVER AEAKAKGKKE DENEDTTEKE EDENEESSQT 660
661 DVGIAKITLQ KVKYLDTTSS MTGSLSPCAE LFIDGQKVKS YRTLRRINEP SWNETIEVLV 720
721 PSKSNSKFVL KIFDDRMNGK ALICEYSSSL DDIMTTLDTA QEFVKGSPQG DIYLDVSWKS 780
781 IEMTGAFAAA NSVSEPIGCI KLDVKDAIIK GDLSGVGDVD PYYTVSLNRR VLYKSIYHSD 840
841 TDHPIFDNST YVPIFSPNQI LTLEFHDYQK IGKDRFIGSV QIPTSNVFKK DPKSGKYVGN 900
901 NGKEEISKLK LKDHEHKVTE SIVNVSTTFI PINLVYSPEE LVNVEKLEKE LKEKKKKFEA 960
961 TQEENEQEME KNPKEWEVAE IEDPFDSDEK KINRKAKLSL NELIKQKSGI LSMQILEGTL1020
1021 SPSSAYLEIL ADDISYPVFI CMKPSQGKLN SEMANIFIRD LNYSKLHFRV SKKHIAKDSD1080
1081 DVISETSYST LKLLKQAYEE PMWLNFNGSK MKVRFLYTPT SVKLPSSESV EDTGYLNIKL1140
1141 ISGHGLKSAD RNGYSDPFVH IFVNDKKVFK SNIKKKTLDP VWNEDAKIPI LSRSKNQVIF1200
1201 NVLDWDRAGD NDDLGQASLD VSSLEVGKTY NWNLNLNTQG SIKLQGSFNP EYIKPSFDIV1260
1261 KGGITDKPMK IASGAAHATV GIAGTGIGAA TGVATGGLKK GGHLLKSLGG NPMKRSKSSN1320
1321 GNESNGAKKS SEKKSFDRRS PSNLNSTSVT PRASLDYDPS VPNTSYAPVQ SASPVVKPTD1380
1381 NTSSSSNKKD TPSSNSRGHS RASSFARTLA PHGTYNGFIT VVAAENVAKH VQIKISLTQG1440
1441 GRLKHIYKTK SQKANNDGVA VFDEECSFKA SPEANLVLGA ISHQRLSRDK DLGIAQINLG1500
1501 DPQIQQDGQI SVKLGDGHLI VKINYGKDKN GQVPPVPEVP QEYTQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
FKS1 TCB3 |
| View Details | Krogan NJ, et al. (2006) |
|
CBR1 GSC2 RPL43B, RPL43A TCB3 |
| View Details | Gavin AC, et al. (2006) |

|
RPL1A, RPL1B TCB3 |
| View Details | Gavin AC, et al. (2002) |
|
DRS2 FKS1 FLC2 PDR5 TCB3 |
| View Details | Ho Y, et al. (2002) |
|
ARO1 ARP2 CPA1 PRB1 PRP28 RNQ1 SWM1 TCB3 URA7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample bir 1 from feb 2005 | Sandall S, et all (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
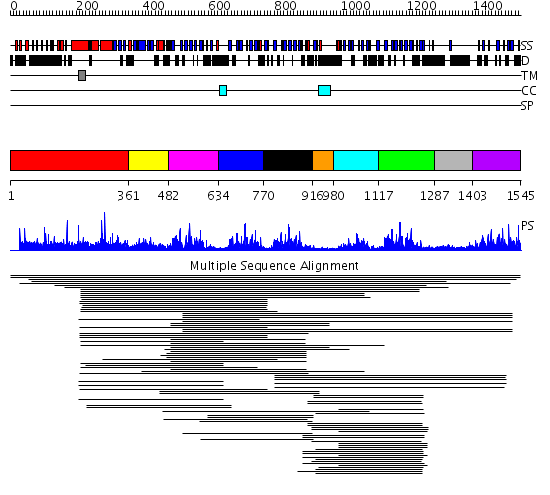
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..360] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [361..481] | 2.136988 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [482..633] | 94.502279 | Synaptogamin I | |
| 4 | View Details | [634..769] | 16.164997 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [770..915] | 65.9897 | C2 domain from protein kinase c (alpha) | |
| 6 | View Details | [916..979] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [980..1116] | 149.503332 | Synaptogamin I | |
| 8 | View Details | [1117..1286] | 149.503332 | Synaptogamin I | |
| 9 | View Details | [1287..1402] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1403..1545] | 10.073995 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
|||||||||
| 5 |
|
|||||||||
| 6 |
|
|||||||||
| 7 | No functions predicted. | |||||||||
| 8 | No functions predicted. | |||||||||
| 9 | No functions predicted. | |||||||||
| 10 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|