













| General Information: |
|
| Name(s) found: |
CPA1 /
YOR303W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 411 amino acids |
Gene Ontology: |
|
| Cellular Component: |
carbamoyl-phosphate synthase complex
[ISS |
| Biological Process: |
arginine biosynthetic process
[TAS |
| Molecular Function: |
carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSAATKATF CIQNGPSFEG ISFGANKSVA GETVFTTSLV GYPESMTDPS YRGQILVFTQ 60
61 PLIGNYGVPS GEARDEYNLL KYFESPHIHV VGIVVAEYAY QYSHWTAVES LAQWCQREGV 120
121 AAITGVDTRE LVQYLREQGS SLGRITLADH DPVPYVNPMK TNLVAQVTTK KPFHVSALPG 180
181 KAKANVALID CGVKENIIRC LVKRGANVTV FPYDYRIQDV ASEFDGIFLS NGPGNPELCQ 240
241 ATISNVRELL NNPVYDCIPI FGICLGHQLL ALASGASTHK LKYGNRAHNI PAMDLTTGQC 300
301 HITSQNHGYA VDPETLPKDQ WKPYFVNLND KSNEGMIHLQ RPIFSTQFHP EAKGGPLDTA 360
361 ILFDKFFDNI EKYQLQSQAK SSISLKVTYS TDKSRLQSIN VTKLAKERVL F |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
CPA1 CPA2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CPA1 CPA2 |
| View Details | Krogan NJ, et al. (2006) |
|
CPA1 CPA2 URA2 |
| View Details | Ho Y, et al. (2002) |
|
ARO1 ARP2 CPA1 PRB1 PRP28 RNQ1 SWM1 TCB3 URA7 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
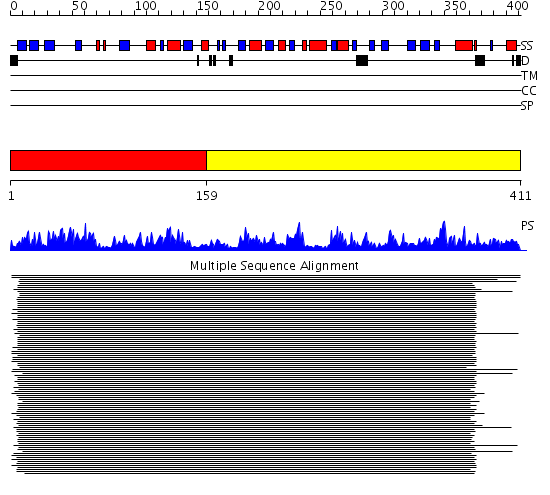
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..158] | 781.0 | Carbamoyl phosphate synthetase, small subunit N-terminal domain; Carbamoyl phosphate synthetase, small subunit C-terminal domain | |
| 2 | View Details | [159..411] | 781.0 | Carbamoyl phosphate synthetase, small subunit N-terminal domain; Carbamoyl phosphate synthetase, small subunit C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)