













| General Information: |
|
| Name(s) found: |
GSC2 /
YGR032W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1895 amino acids |
Gene Ontology: |
|
| Cellular Component: |
prospore membrane
[IDA 1,3-beta-glucan synthase complex [TAS actin cap [IDA |
| Biological Process: |
1,3-beta-glucan biosynthetic process
[TAS cellular cell wall organization [TAS ascospore wall assembly [IGI |
| Molecular Function: |
1,3-beta-glucan synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYNDPNLNG QYYSNGDGTG DGNYPTYQVT QDQSAYDEYG QPIYTQNQLD DGYYDPNEQY 60
61 VDGTQFPQGQ DPSQDQGPYN NDASYYNQPP NMMNPSSQDG ENFSDFSSYG PPSGTYPNDQ 120
121 YTPSQMSYPD QDGSSGASTP YGNGVVNGNG QYYDPNAIEM ALPNDPYPAW TADPQSPLPI 180
181 EQIEDIFIDL TNKFGFQRDS MRNMFDHFMT LLDSRSSRMS PEQALLSLHA DYIGGDTANY 240
241 KKWYFAAQLD MDDEIGFRNM KLGKLSRKAR KAKKKNKKAM QEASPEDTEE TLNQIEGDNS 300
301 LEAADFRWKS KMNQLSPFEM VRQIALFLLC WGEANQVRFT PECLCFIYKC ASDYLDSAQC 360
361 QQRPDPLPEG DFLNRVITPL YRFIRSQVYE IVDGRYVKSE KDHNKVIGYD DVNQLFWYPE 420
421 GIAKIVMEDG TRLIDLPAEE RYLKLGEIPW DDVFFKTYKE TRSWLHLVTN FNRIWIMHIS 480
481 VYWMYCAYNA PTFYTHNYQQ LVDNQPLAAY KWATAALGGT VASLIQVAAT LCEWSFVPRK 540
541 WAGAQHLSRR FWFLCVIMGI NLGPVIFVFA YDKDTVYSTA AHVVGAVMFF VAVATLVFFS 600
601 VMPLGGLFTS YMKKSTRSYV ASQTFTASFA PLHGLDRWMS YLVWVTVFAA KYAESYFFLI 660
661 LSLRDPIRIL STTSMRCTGE YWWGNKICKV QPKIVLGLMI ATDFILFFLD TYLWYIVVNT 720
721 VFSVGKSFYL GISILTPWRN IFTRLPKRIY SKILATTDME IKYKPKVLIS QIWNAIIISM 780
781 YREHLLAIDH VQKLLYHQVP SEIEGKRTLR APTFFVSQDD NNFETEFFPR DSEAERRISF 840
841 FAQSLSTPIP EPLPVDNMPT FTVLTPHYAE RILLSLREII REDDQFSRVT LLEYLKQLHP 900
901 VEWDCFVKDT KILAEETAAY ENNEDEPEKE DALKSQIDDL PFYCIGFKSA APEYTLRTRI 960
961 WASLRSQTLY RTISGFMNYS RAIKLLYRVE NPEIVQMFGG NADGLERELE KMARRKFKFL1020
1021 VSMQRLAKFK PHELENAEFL LRAYPDLQIA YLDEEPPLNE GEEPRIYSAL IDGHCEILEN1080
1081 GRRRPKFRVQ LSGNPILGDG KSDNQNHALI FYRGEYIQLI DANQDNYLEE CLKIRSVLAE1140
1141 FEELGIEQIH PYTPGLKYED QSTNHPVAIV GAREYIFSEN SGVLGDVAAG KEQTFGTLFA1200
1201 RTLAQIGGKL HYGHPDFINA TFMTTRGGVS KAQKGLHLNE DIYAGMNAVL RGGRIKHCEY1260
1261 YQCGKGRDLG FGTILNFTTK IGAGMGEQML SREYYYLGTQ LPIDRFLTFY YAHPGFHLNN1320
1321 LFIQLSLQMF MLTLVNLHAL AHESILCVYD RDKPITDVLY PIGCYNFHPA IDWVRRYTLS1380
1381 IFIVFWIAFV PIVVQELIER GLWKATQRFF RHILSLSPMF EVFAGQIYSS ALLSDIAVGG1440
1441 ARYISTGRGF ATSRIPFSIL YSRFAGSAIY MGSRSMLMLL FGTVAHWQAP LLWFWASLSA1500
1501 LIFAPFIFNP HQFAWEDFFL DYRDYIRWLS RGNNKYHRNS WIGYVRMSRS RVTGFKRKLV1560
1561 GDESEKSAGD ASRAHRTNLI MAEIIPCAIY AAGCFIAFTF INAQTGVKTT DEDRVNSTLR1620
1621 IIICTLAPIV IDIGVLFFCM GLSCCSGPLL GMCCKKTGSV MAGIAHGIAV VVHIVFFIVM1680
1681 WVLEGFSFVR MLIGVVTCIQ CQRLIFHCMT VLLLTREFKN DHANTAFWTG KWYSTGLGYM1740
1741 AWTQPTRELT AKVIELSEFA ADFVLGHVIL IFQLPVICIP KIDKFHSIML FWLKPSRQIR1800
1801 PPIYSLKQAR LRKRMVRRYC SLYFLVLIIF AGCIVGPAVA SAHVPKDLGS GLTGTFHNLV1860
1861 QPRNVSNNDT GSQMSTYKSH YYTHTPSLKT WSTIK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
FEN1 FKS1 GSC2 |
| View Details | Krogan NJ, et al. (2006) |
|
CBR1 GSC2 RPL43B, RPL43A TCB3 |
| View Details | Qiu et al. (2008) |
|
CHS3 FKS1 GSC2 RHO1 SPS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Lockshon D, et al. (2006) |
New Feature: Upload Your Own Microscopy Data
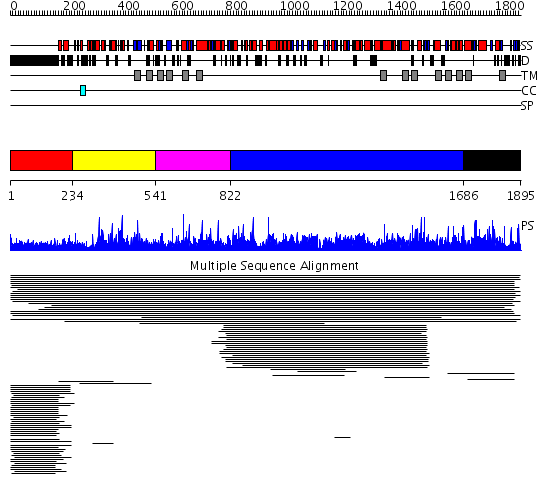
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | 177.324997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [234..540] | 1.03395 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [541..821] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [822..1685] | 1000.0 | 1,3-beta-glucan synthase component No confident structure predictions are available. | |
| 5 | View Details | [1686..1895] | 1.019941 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [870..961] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [962..1123] | 2.052944 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1124..1256] | 3.052946 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1257..1446] | 2.054926 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1447..1685] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1686..1895] | 2.02494 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|