













| General Information: |
|
| Name(s) found: |
YER138C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1755 amino acids |
Gene Ontology: |
|
| Cellular Component: |
retrotransposon nucleocapsid
[ISS |
| Biological Process: |
transposition, RNA-mediated
[ISS |
| Molecular Function: |
DNA-directed DNA polymerase activity
[ISS RNA-directed DNA polymerase activity [ISS RNA binding [ISS protein binding [ISS ribonuclease activity [ISS peptidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESQQLSQHS PISHGSACAS VTSKEVHTNQ DPLDVSASKT EECEKASTKA NSQQTTTPAS 60
61 SAVPENPHHA SPQPASVPPP QNGPYPQQCM MTQNQANPSG WSFYGHPSMI PYTPYQMSPM 120
121 YFPPGPQSQF PQYPSSVGTP LSTPSPESGN TFTDSSSADS DMTSTKKYVR PPPMLTSPND 180
181 FPNWVKTYIK FLQNSNLGGI IPTVNGKPVR QITDDELTFL YNTFQIFAPS QFLPTWVKDI 240
241 LSVDYTDIMK ILSKSIEKMQ SDTQEANDIV TLANLQYNGS TPADAFETKV TNIIDRLNNN 300
301 GIHINNKVAC QLIMRGLSGE YKFLRYTRHR HLNMTVAELF LDIHAIYEEQ QGSRNSKPNY 360
361 RRNLSDEKND SRSYTNTTKP KVIARNPQKT NNSKSKTARA HNVSTSNNSP STDNDSISKS 420
421 TTEPIQLNNK HDLHLGQELT ESTVNHTNHS DDELPGHLLL DSGASRTLIR SAHHIHSASS 480
481 NPDINVVDAQ KRNIPINAIG DLQFHFQDNT KTSIKVLHTP NIAYDLLSLN ELAAVDITAC 540
541 FTKNVLERSD GTVLAPIVKY GDFYWVSKKY LLPSNISVPT INNVHTSEST RKYPYPFIHR 600
601 MLAHANAQTI RYSLKNNTIT YFNESDVDWS SAIDYQCPDC LIGKSTKHRH IKGSRLKYQN 660
661 SYEPFQYLHT DIFGPVHNLP KSAPSYFISF TDETTKFRWV YPLHDRREDS ILDVFTTILA 720
721 FIKNQFQASV LVIQMDRGSE YTNRTLHKFL EKNGITPCYT TTADSRAHGV AERLNRTLLD 780
781 DCRTQLQCSG LPNHLWFSAI EFSTIVRNSL ASPKSKKSAR QHAGLAGLDI STLLPFGQPV 840
841 IVNDHNPNSK IHPRGIPGYA LHPSRNSYGY IIYLPSLKKT VDTTNYVILQ GKESRLDQFN 900
901 YDALTFDEDL NRLTASYQSF IASNEIQQSD DLNIESDHDF QSDIELHPEQ PRNVLSKAVS 960
961 PTDSTPPSTH TEDSKRVSKT NIRAPREVDP NISESNILPS KKRSSTPQIS NIESTGSGGM1020
1021 HKLNVPLLAP MSQSNTHESS HASKSKDFRH SDSYSENETN HTNVPISSTG GTNNKTVPQI1080
1081 SDQETEKRII HRSPSIDASP PENNSSHNIV PIKTPTTVSE QNTEESIIAD LPLPDLPPES1140
1141 PTEFPDPFKE LPPINSHQTN SSLGGIGDSN AYTTINSKKR SLEDNETEIK VSRDTWNTKN1200
1201 MRSLEPPRSK KRIHLIAAVK AVKSIKPIRT TLRYDEAITY NKDIKEKEKY IEAYHKEVNQ1260
1261 LLKMKTWDTD EYYDRKEIDP KRVINSMFIF NKKRDGTHKA RFVARGDIQH PDTYDSGMQS1320
1321 NTVHHYALMT SLSLALDNNY YITQLDISSA YLYADIKEEL YIRPPPHLGM NDKLIRLKKS1380
1381 LYGLKQSGAN WYETIKSYLI KQCGMEEVRG WSCVFKNSQV TICLFVDDMI LFSKDLNANK1440
1441 KIITTLKKQY DTKIINLGES DNEIQYDILG LEIKYQRGKY MKLGMENSLT EKIPKLNVPL1500
1501 NPKGRKLSAP GQPGLYIDQD ELEIDEDEYK EKVHEMQKLI GLASYVGYKF RFDLLYYINT1560
1561 LAQHILFPSR QVLDMTYELI QFMWDTRDKQ LIWHKNKPTE PDNKLVAISD ASYGNQPYYK1620
1621 SQIGNIYLLN GKVIGGKSTK ASLTCTSTTE AEIHAISESV PLLNNLSYLI QELNKKPIIK1680
1681 GLLTDSRSTI SIIKSTNEEK FRNRFFGTKA MRLRDEVSGN NLYVYYIETK KNIADVMTKP1740
1741 LPIKTFKLLT NKWIH |
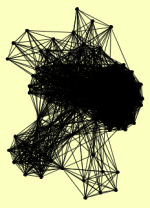
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
AAC3 ABP1 ACC1 ACO1 ADR1 AHA1 AIM41 ATP3 ATP4 BEM2 BUD14 BUR2 CAR2 CDC25 CDC28 CKI1 CKS1 CLB2 CLB3 CLB5 CLN1 COR1 CPR6 CRZ1 CYS4 DCP1 DCP2 DNM1 DOP1 ECM10 ECM29 EDC3 EDE1 EGD2 ENP1 FAA4 GAS1 GCD11 GCN3 GCR2 GIN4 GPH1 HHF1, HHF2 HOM3 HOR2 HRR25 HSP104 HXT7 HYP2 ILV2 IPP1 IRA1 IRA2 KAP122 LIA1 LOC1 LSP1 LTV1 MDH1 MDS3 MET18 MGE1 MYO2 MYO4 NAP1 NSA1 NUG1 OLA1 OYE2 PEX19 PIL1 PIN4 PMD1 PRB1 PRS2 PRS3 PRS5 PTC3 PTC4 PUF3 RAD52 RAD59 RHR2 RIM11 RIO2 RNR2 RPA135 RPA190 RPC19 RPC40 RPM2 RPO31 RPT4 SAP185 SAP190 SAS10 SEC2 SEC23 SEC27 SEC53 SES1 SFB3 SGD1 SGM1 SIT4 SRY1 TEF4 TMA19 TPS1 TSL1 TSR1 UBA1 UTP5 VMA4 VMA8 YBR225W YDR170W-A YER138C YER160C YJR027W YJR028W YPT31 |
| View Details | Qiu et al. (2008) |
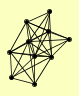
|
YBR012W-B YDR210C-C YDR210C-D YER138C YHR214C-B YJL113W YML045W YOR142W-B YOR343W-A YOR343W-B |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)