













| General Information: |
|
| Name(s) found: |
PHO84 /
YML123C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to plasma membrane
[IDA |
| Biological Process: |
phosphate transport
[IDA manganese ion transport [IMP |
| Molecular Function: |
inorganic phosphate transmembrane transporter activity
[IDA manganese ion transmembrane transporter activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVNKDTIH VAERSLHKEH LTEGGNMAFH NHLNDFAHIE DPLERRRLAL ESIDDEGFGW 60
61 QQVKTISIAG VGFLTDSYDI FAINLGITMM SYVYWHGSMP GPSQTLLKVS TSVGTVIGQF 120
121 GFGTLADIVG RKRIYGMELI IMIVCTILQT TVAHSPAINF VAVLTFYRIV MGIGIGGDYP 180
181 LSSIITSEFA TTKWRGAIMG AVFANQAWGQ ISGGIIALIL VAAYKGELEY ANSGAECDAR 240
241 CQKACDQMWR ILIGLGTVLG LACLYFRLTI PESPRYQLDV NAKLELAAAA QEQDGEKKIH 300
301 DTSDEDMAIN GLERASTAVE SLDNHPPKAS FKDFCRHFGQ WKYGKILLGT AGSWFTLDVA 360
361 FYGLSLNSAV ILQTIGYAGS KNVYKKLYDT AVGNLILICA GSLPGYWVSV FTVDIIGRKP 420
421 IQLAGFIILT ALFCVIGFAY HKLGDHGLLA LYVICQFFQN FGPNTTTFIV PGECFPTRYR 480
481 STAHGISAAS GKVGAIIAQT ALGTLIDHNC ARDGKPTNCW LPHVMEIFAL FMLLGIFTTL 540
541 LIPETKRKTL EEINELYHDE IDPATLNFRN KNNDIESSSP SQLQHEA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
FET4 PHO84 |
| View Details | Ho Y, et al. (2002) |

|
AHA1 ARP2 ARP7 ART10 ATP3 BRX1 BUL1 BZZ1 CKB1 COX6 CPR6 DED1 DIG1 DIG2 DMA2 DUN1 ECM29 FAR10 FAR7 FAR8 FET4 GAL2 GAR1 GDE1 GFA1 GUS1 GYP5 HAS1 HOM6 HXT6 HXT7 IDH1 ILV5 KSS1 LAS17 LSB1 LSP1 LYS12 MET18 MKT1 MSE1 NAP1 NIP7 NMD3 NMD5 NOC2 NOP1 NOP2 NOP56 NOP7 NSA1 NSA2 NUG1 PEP1 PET10 PHO84 PIL1 PIM1 PMA1 PPH21 PPH22 PRB1 PRE10 PYC1 REP1 RGC1 RNQ1 RPA135 RPB3 RPB5 RPN1 RPN10 RPN12 RPN8 RPO21 RPO26 RRP1 RSP5 RVS167 SDH2 SEN1 SLA1 SPS1 SQT1 STE12 STE7 STH1 TEC1 UBI4 UBP7 URA7 VMA6 VMA8 VPS64 VPS74 YDR239C YHM2 YHR033W YKR018C YLR243W YPL009C |
| View Details | Qiu et al. (2008) |
|
ARR3 ATP6 ATP8 CTR3 ENB1 GAP1 OLI1 PHO84 PHO89 PMR1 SMF1 SSU1 SUL2 ZRT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | sample: hx1 | Hao Xu, et al. (2010) | |
| View Run | sample: hx2 | Hao Xu, et al. (2010) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
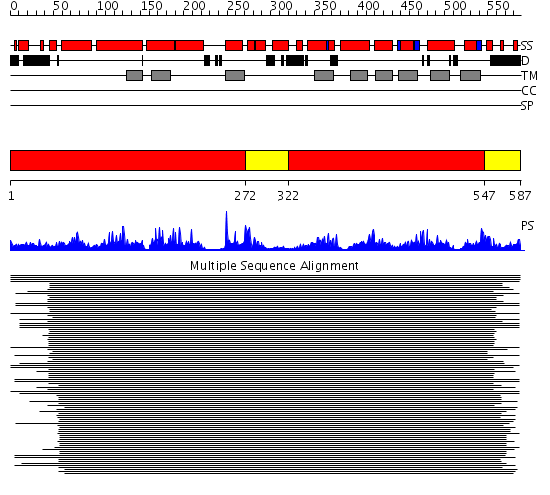
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..271] [322..546] |
137.9897 | No description for 1ja5A_ was found. | |
| 2 | View Details | [272..321] [547..587] |
137.9897 | No description for 1ja5A_ was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|