













| General Information: |
|
| Name(s) found: |
RSP5 /
YER125W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[IDA cytoplasm [IDA mitochondrion [IDA Golgi apparatus [IDA endosome membrane [IDA plasma membrane [IDA |
| Biological Process: |
mRNA export from nucleus
[IMP farnesyl diphosphate biosynthetic process, mevalonate pathway [IMP transcription [TAS protein polyubiquitination [IDA chromatin assembly or disassembly [IMP endocytosis [TAS protein monoubiquitination [IDA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSSISVKLV AAESLYKRDV FRSPDPFAVL TIDGYQTKST SAAKKTLNPY WNETFKFDDI 60
61 NENSILTIQV FDQKKFKKKD QGFLGVVNVR VGDVLGHLDE DTATSSGRPR EETITRDLKK 120
121 SNDGMAVSGR LIVVLSKLPS SSPHSQAPSG HTASSSTNTS STTRTNGHST SSTRNHSTSH 180
181 PSRGTAQAVE STLQSGTTAA TNTATTSHRS TNSTSSATRQ YSSFEDQYGR LPPGWERRTD 240
241 NFGRTYYVDH NTRTTTWKRP TLDQTEAERG NQLNANTELE RRQHRGRTLP GGSSDNSSVT 300
301 VQVGGGSNIP PVNGAAAAAF AATGGTTSGL GELPSGWEQR FTPEGRAYFV DHNTRTTTWV 360
361 DPRRQQYIRT YGPTNTTIQQ QPVSQLGPLP SGWEMRLTNT ARVYFVDHNT KTTTWDDPRL 420
421 PSSLDQNVPQ YKRDFRRKVI YFRSQPALRI LPGQCHIKVR RKNIFEDAYQ EIMRQTPEDL 480
481 KKRLMIKFDG EEGLDYGGVS REFFFLLSHE MFNPFYCLFE YSAYDNYTIQ INPNSGINPE 540
541 HLNYFKFIGR VVGLGVFHRR FLDAFFVGAL YKMMLRKKVV LQDMEGVDAE VYNSLNWMLE 600
601 NSIDGVLDLT FSADDERFGE VVTVDLKPDG RNIEVTDGNK KEYVELYTQW RIVDRVQEQF 660
661 KAFMDGFNEL IPEDLVTVFD ERELELLIGG IAEIDIEDWK KHTDYRGYQE SDEVIQWFWK 720
721 CVSEWDNEQR ARLLQFTTGT SRIPVNGFKD LQGSDGPRRF TIEKAGEVQQ LPKSHTCFNR 780
781 VDLPQYVDYD SMKQKLTLAV EETIGFGQE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
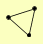
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
RSP5 RUP1 UBP2 |
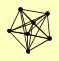
| View Details | Krogan NJ, et al. (2006) |
|
ADE6 BUL1 LDB19 RSP5 RUP1 UBP2 YHR131C |
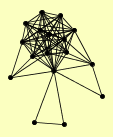
| View Details | Ho Y, et al. (2002) |
|
ART10 BUL1 DUN1 HXT6 LSB1 MMS2 PHO84 RNQ1 RPB3 RPB5 RPO21 RPO26 RSP5 RVS161 RVS167 UBC13 YKR018C |
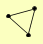
| View Details | Qiu et al. (2008) |
|
APC9 CDC16 RSP5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
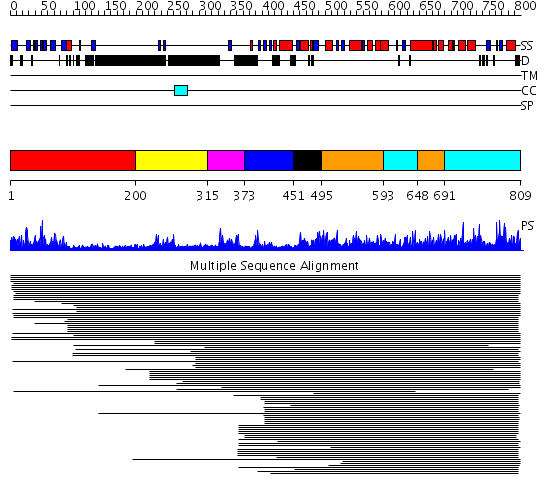
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 31.69897 | Synaptogamin I | |
| 2 | View Details | [200..314] | 37.67837 | Mitotic rotamase PIN1; Mitotic rotamase PIN1, domain 2 | |
| 3 | View Details | [315..372] | 70.264245 | Ubiquitin ligase NEDD4 WWIII domain | |
| 4 | View Details | [373..450] | 10.59 | Ubiquitin ligase NEDD4 WWIII domain | |
| 5 | View Details | [451..494] | 664.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 6 | View Details | [495..592] [648..690] |
664.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 7 | View Details | [593..647] [691..809] |
664.0 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)