













| General Information: |
|
| Name(s) found: |
PUF3 /
YLL013C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 879 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA external side of mitochondrial outer membrane [IDA |
| Biological Process: |
mRNA catabolic process
[IDA aerobic respiration [IMP mitochondrion localization [IMP mitochondrion organization [IMP nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [IMP |
| Molecular Function: |
mRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEMNMDMDMD MELASIVSSL SALSHSNNNG GQAAAAGIVN GGAAGSQQIG GFRRSSFTTA 60
61 NEVDSEILLL HGSSESSPIF KKTALSVGTA PPFSTNSKKF FGNGGNYYQY RSTDTASLSS 120
121 ASYNNYHTHH TAANLGKNNK VNHLLGQYSA SIAGPVYYNG NDNNNSGGEG FFEKFGKSLI 180
181 DGTRELESQD RPDAVNTQSQ FISKSVSNAS LDTQNTFEQN VESDKNFNKL NRNTTNSGSL 240
241 YHSSSNSGSS ASLESENAHY PKRNIWNVAN TPVFRPSNNP AAVGATNVAL PNQQDGPANN 300
301 NFPPYMNGFP PNQFHQGPHY QNFPNYLIGS PSNFISQMIS VQIPANEDTE DSNGKKKKKA 360
361 NRPSSVSSPS SPPNNSPFPF AYPNPMMFMP PPPLSAPQQQ QQQQQQQQQE DQQQQQQQEN 420
421 PYIYYPTPNP IPVKMPKDEK TFKKRNNKNH PANNSNNANK QANPYLENSI PTKNTSKKNA 480
481 SSKSNESTAN NHKSHSHSHP HSQSLQQQQQ TYHRSPLLEQ LRNSSSDKNS NSNMSLKDIF 540
541 GHSLEFCKDQ HGSRFIQREL ATSPASEKEV IFNEIRDDAI ELSNDVFGNY VIQKFFEFGS 600
601 KIQKNTLVDQ FKGNMKQLSL QMYACRVIQK ALEYIDSNQR IELVLELSDS VLQMIKDQNG 660
661 NHVIQKAIET IPIEKLPFIL SSLTGHIYHL STHSYGCRVI QRLLEFGSSE DQESILNELK 720
721 DFIPYLIQDQ YGNYVIQYVL QQDQFTNKEM VDIKQEIIET VANNVVEYSK HKFASNVVEK 780
781 SILYGSKNQK DLIISKILPR DKNHALNLED DSPMILMIKD QFANYVIQKL VNVSEGEGKK 840
841 LIVIAIRAYL DKLNKSNSLG NRHLASVEKL AALVENAEV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
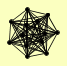
|
BNI4 BUD14 GIP3 GLC7 GLC8 HER1 PPZ1 PUF3 SDS22 SOL1 SOL2 YLR257W YPI1 |
| View Details | Gavin AC, et al. (2002) |
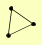
|
LSM12 PUF3 TUB3 |
| View Details | Ho Y, et al. (2002) |

|
ABP1 ACO1 AIM41 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 ATP3 ATP4 AVO1 BUD14 CAR2 CCT5 CLU1 COR1 CRZ1 CYS4 DCP1 DCP2 DHH1 DNM1 DPB2 EDC3 EDE1 EGD2 ENP1 FET3 FUN12 GAL7 GAS1 GCN3 GFA1 GPH1 HHF1, HHF2 HRR25 HSP104 HXT7 HYP2 IDH1 ILV2 IPP1 LOC1 LSP1 LTV1 MDH1 MDS3 MET16 MGE1 MKK2 NOG2 NPA3 NSA1 NUG1 OYE2 PDX1 PEX19 PHO85 PIL1 PIN4 PST2 PTC4 PUF3 RIO2 RPC19 RPM2 RPN12 RPN8 RVB1 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SIS1 SIT4 SLX1 SRP54 STI1 TEM1 TFC7 TMA19 TSR1 VMA4 VMA8 YBR225W YER077C YER138C YGR266W YKR051W YKU80 YLR271W YPR003C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
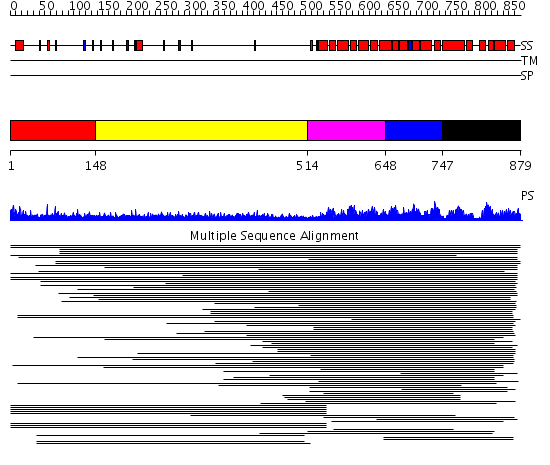
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 3.404997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [148..513] | 14.59 | RBP1 | |
| 3 | View Details | [514..647] | 973.9794 | Pumilio 1 | |
| 4 | View Details | [648..746] | 973.9794 | Pumilio 1 | |
| 5 | View Details | [747..879] | 973.9794 | Pumilio 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)