













| General Information: |
|
| Name(s) found: |
GUD1 /
YDL238C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 489 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
guanine metabolic process
[ISS |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides
[ISS guanine deaminase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKSDLLFDK FNDKHGKFLV FFGTFVDTPK LGELRIREKT SVGVLNGIIR FVNRNSLDPV 60
61 KDCLDHDSSL SPEDVTVVDI IGKDKTRNNS FYFPGFVDTH NHVSQYPNVG VFGNSTLLDW 120
121 LEKYTFPIEA ALANENIARE VYNKVISKTL SHGTTTVAYY NTIDLKSTKL LAQLSSLLGQ 180
181 RVLVGKVCMD TNGPEYYIED TKTSFESTVK VVKYIRETIC DPLVNPIVTP RFAPSCSREL 240
241 MQQLSKLVKD ENIHVQTHLS ENKEEIQWVQ DLFPECESYT DVYDKYGLLT EKTVLAHCIH 300
301 LTDAEARVIK QRRCGISHCP ISNSSLTSGE CRVRWLLDQG IKVGLGTDVS AGHSCSILTT 360
361 GRQAFAVSRH LAMRETDHAK LSVSECLFLA TMGGAQVLRM DETLGTFDVG KQFDAQMIDT 420
421 NAPGSNVDMF HWQLKEKDQM QEQEQEQGQD PYKNPPLLTN EDIIAKWFFN GDDRNTTKVW 480
481 VAGQQVYQI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AAP1 GUD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
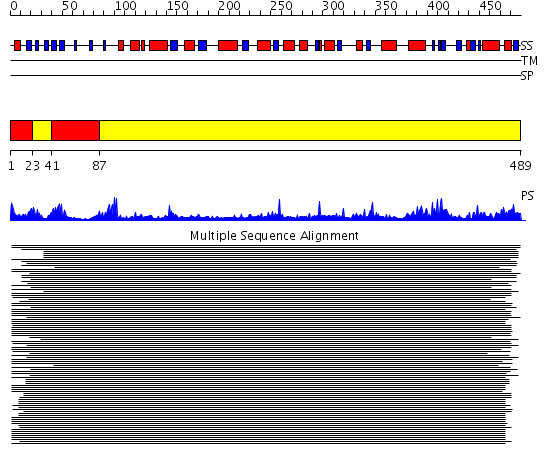
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..22] [41..86] |
64.69897 | alpha-Subunit of urease; alpha-subunit of urease, catalytic domain | |
| 2 | View Details | [23..40] [87..489] |
64.69897 | alpha-Subunit of urease; alpha-subunit of urease, catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)