













| General Information: |
|
| Name(s) found: |
JSN1 /
YJR091C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1091 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay
[IGI |
| Molecular Function: |
mRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKSKQMNIN NLSNIPEVID PGITIPIYEE EYENNGESNS QLQQQPQKLG SYRSRAGKFS 60
61 NTLSNLLPSI SAKLHHSKKN SHGKNGAEFS SSNNSSQSTV ASKTPRASPS RSKMMESSID 120
121 GVTMDRPGSL TPPQDMEKLV HFPDSSNNFL IPAPRGSSDS FNLPHQISRT RNNTMSSQIT 180
181 SISSIAPKPR TSSGIWSSNA SANDPMQQHL LQQLQPTTSN NTTNSNTLND YSTKTAYFDN 240
241 MVSTSGSQMA DNKMNTNNLA IPNSVWSNTR QRSQSNASSI YTDAPLYEQP ARASISSHYT 300
301 IPTQESPLIA DEIDPQSINW VTMDPTVPSI NQISNLLPTN TISISNVFPL QHQQPQLNNA 360
361 INLTSTSLAT LCSKYGEVIS ARTLRNLNMA LVEFSSVESA VKALDSLQGK EVSMIGAPSK 420
421 ISFAKILPMH QQPPQFLLNS QGLPLGLENN NLQPQPLLQE QLFNGAVTFQ QQGNVSIPVF 480
481 NQQSQQSQHQ NHSSGSAGFS NVLHGYNNNN SMHGNNNNSA NEKEQCPFPL PPPNVNEKED 540
541 LLREIIELFE ANSDEYQINS LIKKSLNHKG TSDTQNFGPL PEPLSGREFD PPKLRELRKS 600
601 IDSNAFSDLE IEQLAIAMLD ELPELSSDYL GNTIVQKLFE HSSDIIKDIM LRKTSKYLTS 660
661 MGVHKNGTWA CQKMITMAHT PRQIMQVTQG VKDYCTPLIN DQFGNYVIQC VLKFGFPWNQ 720
721 FIFESIIANF WVIVQNRYGA RAVRACLEAH DIVTPEQSIV LSAMIVTYAE YLSTNSNGAL 780
781 LVTWFLDTSV LPNRHSILAP RLTKRIVELC GHRLASLTIL KVLNYRGDDN ARKIILDSLF 840
841 GNVNAHDSSP PKELTKLLCE TNYGPTFVHK VLAMPLLEDD LRAHIIKQVR KVLTDSTQIQ 900
901 PSRRLLEEVG LASPSSTHNK TKQQQQQHHN SSISHMFATP DTSGQHMRGL SVSSVKSGGS 960
961 KHTTMNTTTT NGSSASTLSP GQPLNANSNS SMGYFSYPGV FPVSGFSGNA SNGYAMNNDD1020
1021 LSSQFDMLNF NNGTRLSLPQ LSLTNHNNTT MELVNNVGSS QPHTNNNNNN NNTNYNDDNT1080
1081 VFETLTLHSA N |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
IRC3 JSN1 RBL2 YJL107C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
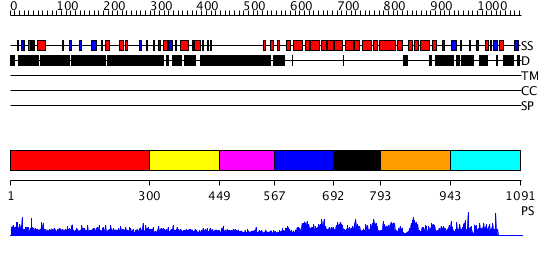
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 17.484997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [300..448] | 13.69 | No description for 1h2tZ was found. | |
| 3 | View Details | [449..566] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [567..691] | 185.0 | Pumilio 1 | |
| 5 | View Details | [692..792] | 185.0 | Pumilio 1 | |
| 6 | View Details | [793..942] | 185.0 | Pumilio 1 | |
| 7 | View Details | [943..1091] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)