













| General Information: |
|
| Name(s) found: |
FUN12 /
YAL035W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1002 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
translational initiation
[IDA |
| Molecular Function: |
translation initiation factor activity
[IDA GTPase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKSKKNQQ NYWDEEFEED AAQNEEISAT PTPNPESSAG ADDTSREASA SAEGAEAIEG 60
61 DFMSTLKQSK KKQEKKVIEE KKDGKPILKS KKEKEKEKKE KEKQKKKEQA ARKKAQQQAQ 120
121 KEKNKELNKQ NVEKAAAEKA AAEKSQKSKG ESDKPSASAK KPAKKVPAGL AALRRQLELK 180
181 KQLEEQEKLE REEEERLEKE EEERLANEEK MKEEAKAAKK EKEKAKREKR KAEGKLLTRK 240
241 QKEEKKLLER RRAALLSSGN VKVAGLAKKD GEENKPKKVV YSKKKKRTTQ ENASEAIKSD 300
301 SKKDSEVVPD DELKESEDVL IDDWENLALG DDDEEGTNEE TQESTASHEN EDQNQGEEEE 360
361 EGEEEEEEEE ERAHVHEVAK STPAATPAAT PTPSSASPNK KDLRSPICCI LGHVDTGKTK 420
421 LLDKIRQTNV QGGEAGGITQ QIGATYFPID AIKAKTKVMA EYEKQTFDVP GLLVIDTPGH 480
481 ESFSNLRSRG SSLCNIAILV IDIMHGLEQQ TIESIKLLRD RKAPFVVALN KIDRLYDWKA 540
541 IPNNSFRDSF AKQSRAVQEE FQSRYSKIQL ELAEQGLNSE LYFQNKNMSK YVSIVPTSAV 600
601 TGEGVPDLLW LLLELTQKRM SKQLMYLSHV EATILEVKVV EGFGTTIDVI LSNGYLREGD 660
661 RIVLCGMNGP IVTNIRALLT PQPLRELRLK SEYVHHKEVK AALGVKIAAN DLEKAVSGSR 720
721 LLVVGPEDDE DELMDDVMDD LTGLLDSVDT TGKGVVVQAS TLGSLEALLD FLKDMKIPVM 780
781 SIGLGPVYKR DVMKASTMLE KAPEYAVMLC FDVKVDKEAE QYAEQEGIKI FNADVIYHLF 840
841 DSFTAYQEKL LEERRKDFLD YAIFPCVLQT LQIINKRGPM IIGVDVLEGT LRVGTPICAV 900
901 KTDPTTKERQ TLILGKVISL EINHQPVQEV KKGQTAAGVA VRLEDPSGQQ PIWGRHVDEN 960
961 DTLYSLVSRR SIDTLKDKAF RDQVARSDWL LLKKLKVVFG IE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
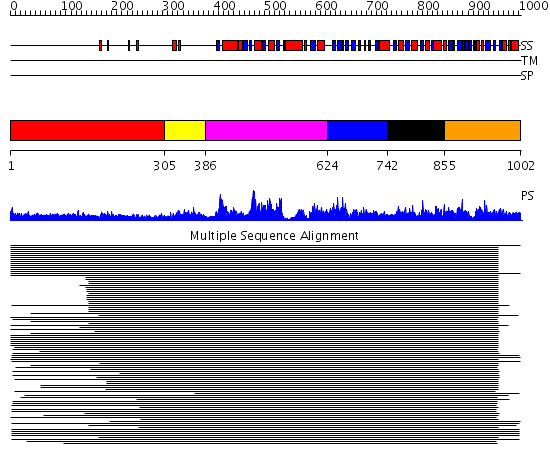
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..304] | 3.09691 | Heavy meromyosin subfragment | |
| 2 | View Details | [305..385] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [386..623] | 1064.0 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain | |
| 4 | View Details | [624..741] | 1064.0 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain | |
| 5 | View Details | [742..854] | 1064.0 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain | |
| 6 | View Details | [855..1002] | 1064.0 | Initiation factor IF2/eIF5b, domains 2 and 4; Initiation factor IF2/eIF5b, domain 3; Initiation factor IF2/eIF5b, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)