













| Protein: | TIF4632 |
| From Publication: | Gavin A.C. et al. (2002) Abstract Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002 Jan 10;415(6868):141-7. |
| Complexes containing TIF4632: | 7 |
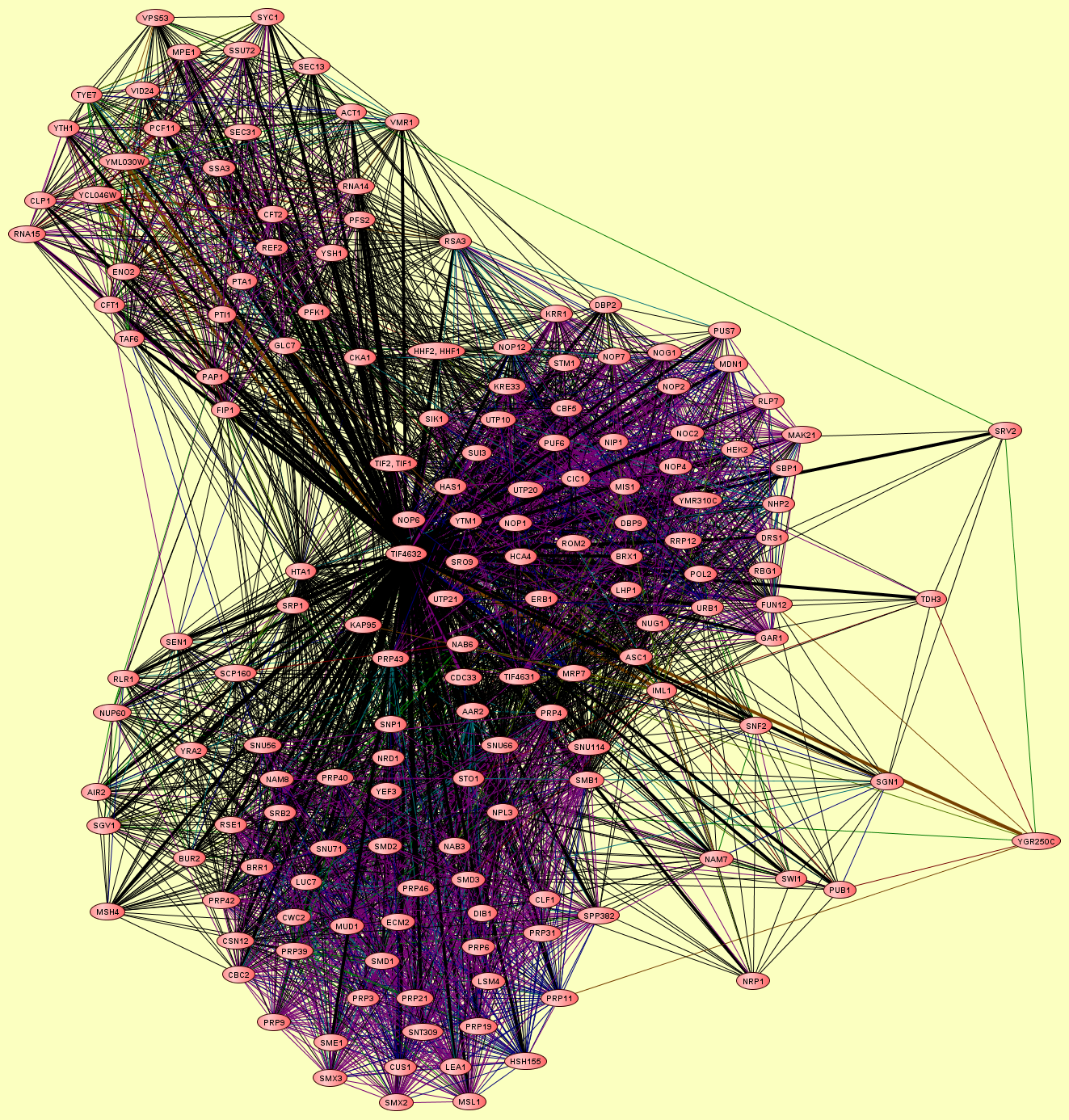
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||