













| General Information: |
|
| Name(s) found: |
SSF1 /
YHR066W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 453 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
conjugation with cellular fusion
[IGI regulation of cell size [IMP ribosomal large subunit assembly [IMP |
| Molecular Function: |
rRNA binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRRQKKRT HAQLTPEQEQ GIPKSMVIRV GQTSLANHSL NQLVKDFRQI MQPHTAIKLK 60
61 ERKSNKLKDF VVMCGPLGVT HLFMFTQSEK TGNVSLKIAR TPQGPTVTFQ VLDYSLGRDI 120
121 KKFLKRPKSL NNDDVLNPPL LVLNGFSTSK RSGEDDQDVN VEKVIVSMFQ NIFPPLNPAR 180
181 TSLNSIKRVF MINKDRETGE ISMRHYFIDI REVEISRNLK RLYKAKNNLS KTVPNLHRKE 240
241 DISSLILDHD LGAYTSESEI EDDAIVRVVD NQDVKAKHSQ SLKSQRTPVE KKDNKEREKE 300
301 TEEEDVEMEE PKPSENLQPT PRKKAIKLTE LGPRLTLKLV KIEEGICSGK VLHHEFVQKS 360
361 SEEIKALEKR HAAKMRLKEQ RKKEQEENIA KKKAVKDAKK QRKLERRKAR AAEGGEGQGK 420
421 DDAMSDDESS SSDSEHYGSV PEDLDSDLFS EVE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
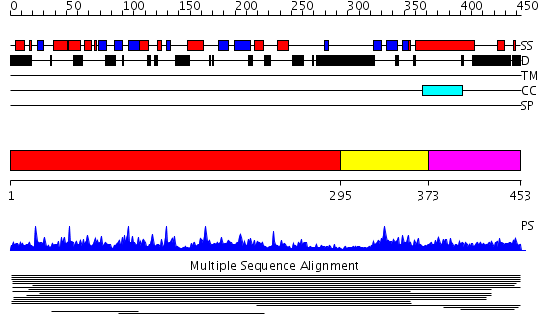
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..294] | 172.79588 | No description for PF03877 was found. No confident structure predictions are available. | |
| 2 | View Details | [295..372] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [373..453] | 2.01199 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.70 |
Source: Reynolds et al. (2008)