













| General Information: |
|
| Name(s) found: |
HDA1 /
YNL021W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 706 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[IDA |
| Biological Process: |
loss of chromatin silencing involved in replicative cell aging
[IMP regulation of transcription, DNA-dependent [TAS protein amino acid deacetylation [IDA chromatin organization [TAS |
| Molecular Function: |
histone deacetylase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSVMVKKEV LENPDHDLKR KLEENKEEEN SLSTTSKSKR QVIVPVCMPK IHYSPLKTGL 60
61 CYDVRMRYHA KIFTSYFEYI DPHPEDPRRI YRIYKILAEN GLINDPTLSG VDDLGDLMLK 120
121 IPVRAATSEE ILEVHTKEHL EFIESTEKMS REELLKETEK GDSVYFNNDS YASARLPCGG 180
181 AIEACKAVVE GRVKNSLAVV RPPGHHAEPQ AAGGFCLFSN VAVAAKNILK NYPESVRRIM 240
241 ILDWDIHHGN GTQKSFYQDD QVLYVSLHRF EMGKYYPGTI QGQYDQTGEG KGEGFNCNIT 300
301 WPVGGVGDAE YMWAFEQVVM PMGREFKPDL VIISSGFDAA DGDTIGQCHV TPSCYGHMTH 360
361 MLKSLARGNL CVVLEGGYNL DAIARSALSV AKVLIGEPPD ELPDPLSDPK PEVIEMIDKV 420
421 IRLQSKYWNC FRRRHANSGC NFNEPINDSI ISKNFPLQKA IRQQQQHYLS DEFNFVTLPL 480
481 VSMDLPDNTV LCTPNISESN TIIIVVHDTS DIWAKRNVIS GTIDLSSSVI IDNSLDFIKW 540
541 GLDRKYGIID VNIPLTLFEP DNYSGMITSQ EVLIYLWDNY IKYFPSVAKI AFIGIGDSYS 600
601 GIVHLLGHRD TRAVTKTVIN FLGDKQLKPL VPLVDETLSE WYFKNSLIFS NNSHQCWKEN 660
661 ESRKPRKKFG RVLRCDTDGL NNIIEERFEE ATDFILDSFE EWSDEE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
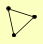
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
HDA1 HDA2 HDA3 |
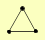
| View Details | Krogan NJ, et al. (2006) |
|
HDA1 HDA2 HDA3 |
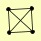
| View Details | Gavin AC, et al. (2006) |
|
HDA1 HDA2 HDA3 PRO2 |
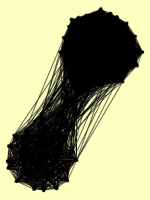
| View Details | Gavin AC, et al. (2002) |
|
ARX1 BEM2 BMS1 BRX1 BUD20 CIC1 CLU1 CMR1 CYR1 CYS4 DBP10 DBP9 DCP2 DNA2 DRS1 EBP2 ERB1 FPR4 GCN1 HAS1 HDA1 HDA2 HDA3 HER1 HIS3 HSC82 INP2 IPI1 IPI3 KAP95 KRE33 LCD1 LHP1 LOC1 LSG1 MAK21 MAK5 MDN1 MEC1 MGM101 MIS1 MLH1 MPH1 MRT4 MSH2 MSH3 MSH6 NAP1 NIP1 NIP7 NMD3 NOC2 NOC3 NOG1 NOG2 NOP1 NOP12 NOP15 NOP16 NOP2 NOP4 NOP56 NOP7 NSA1 NSA2 NUG1 OLA1 PGK1 PMS1 PRO2 PRT1 PUF6 RAD16 RAD52 REI1 RFA1 RFA2 RFA3 RIM1 RIX1 RIX7 RLP24 RLP7 RNH1 RPA135 RPA190 RPF1 RPF2 RPG1 RPP2B RPT6 RRP1 RRP12 RSA3 RSA4 RSC8 RTR1 RVB2 SDA1 SEC7 SGS1 SHS1 SKI2 SNU13 SPB1 SPB4 SSF1 TIF1, TIF2 TIF6 TOP3 TUB3 URB1 UTP10 UTP20 UTP22 VAS1 VPS1 YGL036W YHR122W YTM1 |
| View Details | Qiu et al. (2008) |
|
HDA1 HDA2 HDA3 HOS1 HOS4 HST1 PHO23 RPD3 SAP30 SDS3 SET3 SIN3 SNT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
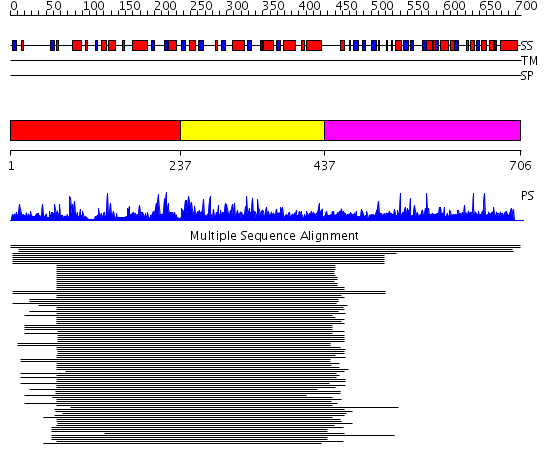
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..236] | 288.522879 | HDAC homologue | |
| 2 | View Details | [237..436] | 288.522879 | HDAC homologue | |
| 3 | View Details | [437..706] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)