













| General Information: |
|
| Name(s) found: |
ADH1 /
YOL086C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 348 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
|
| Biological Process: |
fermentation
[IMP]
NADH oxidation [IDA ethanol biosynthetic process involved in glucose fermentation to ethanol [IMP |
| Molecular Function: |
alcohol dehydrogenase (NAD) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIPETQKGV IFYESHGKLE YKDIPVPKPK ANELLINVKY SGVCHTDLHA WHGDWPLPVK 60
61 LPLVGGHEGA GVVVGMGENV KGWKIGDYAG IKWLNGSCMA CEYCELGNES NCPHADLSGY 120
121 THDGSFQQYA TADAVQAAHI PQGTDLAQVA PILCAGITVY KALKSANLMA GHWVAISGAA 180
181 GGLGSLAVQY AKAMGYRVLG IDGGEGKEEL FRSIGGEVFI DFTKEKDIVG AVLKATDGGA 240
241 HGVINVSVSE AAIEASTRYV RANGTTVLVG MPAGAKCCSD VFNQVVKSIS IVGSYVGNRA 300
301 DTREALDFFA RGLVKSPIKV VGLSTLPEIY EKMEKGQIVG RYVVDTSK |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
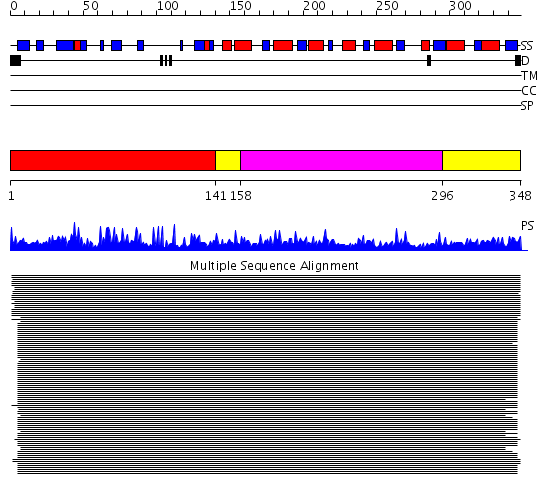
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | 289.0 | Alcohol dehydrogenase | |
| 2 | View Details | [141..157] [296..348] |
289.0 | Alcohol dehydrogenase | |
| 3 | View Details | [158..295] | 289.0 | Alcohol dehydrogenase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)