













| General Information: |
|
| Name(s) found: |
GUS1 /
YGL245W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA methionyl glutamyl tRNA synthetase complex [IDA |
| Biological Process: |
glutamyl-tRNA aminoacylation
[IDA |
| Molecular Function: |
glutamate-tRNA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSTLTINGK APIVAYAELI AARIVNALAP NSIAIKLVDD KKAPAAKLDD ATEDVFNKIT 60
61 SKFAAIFDNG DKEQVAKWVN LAQKELVIKN FAKLSQSLET LDSQLNLRTF ILGGLKYSAA 120
121 DVACWGALRS NGMCGSIIKN KVDVNVSRWY TLLEMDPIFG EAHDFLSKSL LELKKSANVG 180
181 KKKETHKANF EIDLPDAKMG EVVTRFPPEP SGYLHIGHAK AALLNQYFAQ AYKGKLIIRF 240
241 DDTNPSKEKE EFQDSILEDL DLLGIKGDRI TYSSDYFQEM YDYCVQMIKD GKAYCDDTPT 300
301 EKMREERMDG VASARRDRSV EENLRIFTEE MKNGTEEGLK NCVRAKIDYK ALNKTLRDPV 360
361 IYRCNLTPHH RTGSTWKIYP TYDFCVPIVD AIEGVTHALR TIEYRDRNAQ YDWMLQALRL 420
421 RKVHIWDFAR INFVRTLLSK RKLQWMVDKD LVGNWDDPRF PTVRGVRRRG MTVEGLRNFV 480
481 LSQGPSRNVI NLEWNLIWAF NKKVIDPIAP RHTAIVNPVK IHLEGSEAPQ EPKIEMKPKH 540
541 KKNPAVGEKK VIYYKDIVVD KDDADVINVD EEVTLMDWGN VIITKKNDDG SMVAKLNLEG 600
601 DFKKTKHKLT WLADTKDVVP VDLVDFDHLI TKDRLEEDES FEDFLTPQTE FHTDAIADLN 660
661 VKDMKIGDII QFERKGYYRL DALPKDGKPY VFFTIPDGKS VNKYGAKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
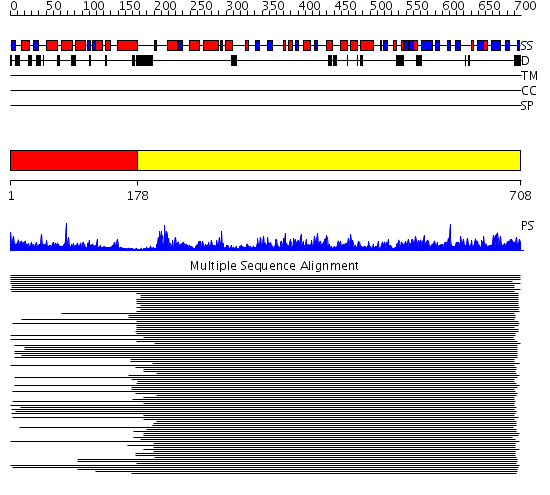
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..177] | 8.30103 | Crystal structure of an insect delta-class glutathione S-transferase from a DDT-resistant strain of the malaria vector Anopheles gambiae | |
| 2 | View Details | [178..708] | 150.0 | Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain; Glutaminyl-tRNA synthetase (GlnRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)