













| General Information: |
|
| Name(s) found: |
RGD1 /
YBR260C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 666 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud
[IDA actin cortical patch [IDA |
| Biological Process: |
response to acid
[IMP osmosensory signaling pathway [IGI |
| Molecular Function: |
Rho GTPase activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEETAKKPAS ATVSAKSSHD GGTDDLAHLF STPEIKKVLN SDVAINALLS RLKQSLLTCE 60
61 EFMKFIRKKY AFEEEHVQEL SKQYKHFFNI QGSTNSSLKK MIHEVLGFDG KMAQVKQSYI 120
121 TALQKMYSEI SSLLLTMTKL RKSVKENSKR LEKDVSDAIH SAEKAQSRYN SLCQDWDKLR 180
181 MTDPTKTKLT LRGSKTTKEQ EEELLRKIDN ADLEYKQKVD HSNSLRNTFI TKERPRIVQE 240
241 LKDLILEIDT AMTIQLQKYT IWTENLVLNT GVTISPLDST KSMKSFAGSV SNERDLYSFL 300
301 NKYNQTGKHS LLINKNLIPV SYKKHPSMNH GQKNKSPPKF AVDPSRNSIP KRMISTHNES 360
361 PFLSSSSNTA AVPNANLNSA TPSLNTNKQL PPTMASSISS TSNAAGAMSP SSSIVTSDTT 420
421 SSITKTLDPG NNSPQIPEEL INSLDSDRPI SHIQTNNNMP PGVQKNFKTF GVPLESLIEF 480
481 EQDMVPAIVR QCIYVIDKFG LDQEGIYRKS ANVLDVSKLK EEIDKDPANI SMILPSKPHS 540
541 DSDIYLVGSL LKTFFASLPD SVLPKALSSE IKVCLQIEDP TTRKNFMHGL IYNLPDAQYW 600
601 TLRALVFHLK RVLAHEAQNR MNLRALCIIW GPTIAPANPD DANDVNFQIM AMEVLLEVSD 660
661 QAFEPE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
FYV8 RGD1 |
| View Details | Krogan NJ, et al. (2006) |
|
FYV8 RGD1 |
| View Details | Ho Y, et al. (2002) |
|
ARP2 BCK1 BUL1 CDC12 DOG1 DOG2 GUS1 IDH1 IMD4 LYS12 MET6 MKK2 MSK1 NPA3 PRB1 PYC1 RGD1 RNQ1 RPN1 RPN7 RVB1 SEC53 TMA19 VMA22 |
| View Details | Qiu et al. (2008) |

|
LRG1 RGD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
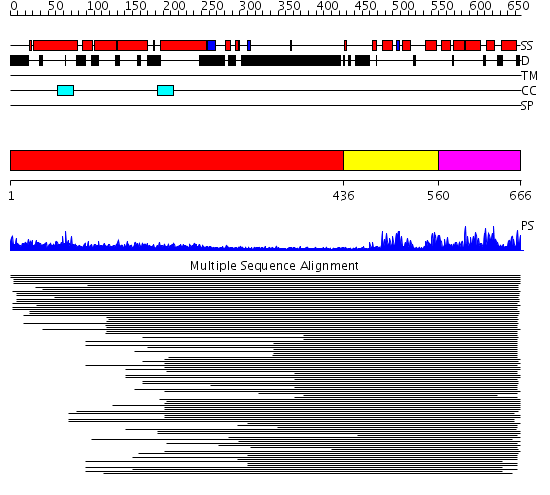
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..435] | 14.39794 | Heavy meromyosin subfragment | |
| 2 | View Details | [436..559] | 166.70927 | p50 RhoGAP domain | |
| 3 | View Details | [560..666] | 166.70927 | p50 RhoGAP domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)