













| General Information: |
|
| Name(s) found: |
TSA2 /
YDR453C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 196 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to oxidative stress
[IGI |
| Molecular Function: |
thioredoxin peroxidase activity
[IDA peroxiredoxin activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAEVQKQAP PFKKTAVVDG IFEEISLEKY KGKYVVLAFV PLAFSFVCPT EIVAFSDAAK 60
61 KFEDQGAQVL FASTDSEYSL LAWTNLPRKD GGLGPVKVPL LADKNHSLSR DYGVLIEKEG 120
121 IALRGLFIID PKGIIRHITI NDLSVGRNVN EALRLVEGFQ WTDKNGTVLP CNWTPGAATI 180
181 KPDVKDSKEY FKNANN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
TSA1 TSA2 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACC1 ACH1 ACO1 ADH2 ADK1 ADR1 AFR1 AHP1 APT1 ARC1 ARF1 ARO1 ATP3 ATP5 ATP7 CAR2 CDC14 CDC33 CDC39 CLU1 COF1 COR1 CPR1 CYR1 CYS4 DCS1 DCS2 DPM1 DSS1 ECM10 EGD1 ERG13 ERG6 FPR1 FRS2 FUR1 GCD11 GDB1 GFA1 GLC7 GND1 GND2 GRX1 GUS1 HEF3 HHF1, HHF2 HMS1 HTB2 HTZ1 IDH2 ISA2 LRO1 LSP1 MCR1 MDH1 MET18 MET6 MTC5 NGL2 NTF2 OLA1 OYE2 PFK1 PIL1 PRE1 PRM2 PST2 PUP3 PYC1 PYC2 RAD16 RAD50 RAD7 RET1 RNR2 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN3 RPO26 RPO31 RSN1 RVS167 SAH1 SCP160 SEC27 SEC53 SEN15 SES1 SHP1 SNF4 SNU13 SOD1 SOD2 SPE3 SSZ1 TBS1 TEF4 TFP1 THS1 TIF1, TIF2 TMA19 TMA29 TPS1 TSA2 UBA1 UBP15 UBR1 VAS1 VMA4 VMA5 VPS1 WTM1 YAK1 YFL042C YHR033W YHR112C YKU80 YLR241W YNK1 YPL247C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
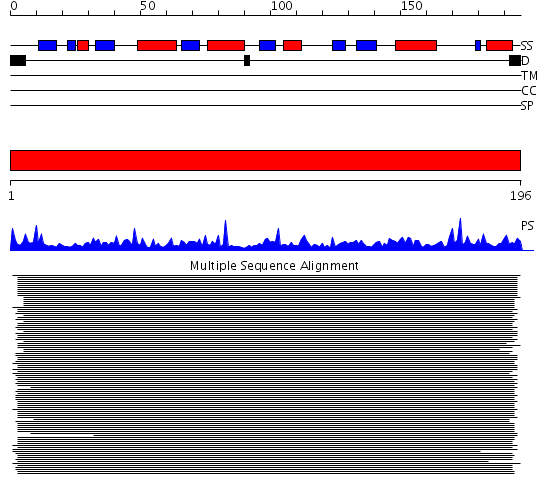
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 733.67837 | Thioredoxin peroxidase 2 (thioredoxin peroxidase B, 2-cys peroxiredoxin) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)