













| General Information: |
|
| Name(s) found: |
DRS1 /
YLL008W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 752 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[TAS nucleolus [TAS |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [TAS |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVGTKKYSN LDFVPTISDS EDDVPILDSS DDEKVEAKKT TKKRKGKNNK KKVSEGDNLD 60
61 EDVHEDLDAG FKFDLDADDT TSNFQGWNFL AEGESNKDDA EAFVKKDVDL DKIIRRKGGL 120
121 VKMAHIDSKQ EEETEKEKVE KENDSDDEEL AMDGFGMGAP MNNGDENQSE EEEEEEEKEE 180
181 EEEEEEEQEE MTLEKGGKDD EIDEEDDSEE AKADFYAPET EGDEAKKQMY ENFNSLSLSR 240
241 PVLKGLASLG YVKPSPIQSA TIPIALLGKD IIAGAVTGSG KTAAFMIPII ERLLYKPAKI 300
301 ASTRVIVLLP TRELAIQVAD VGKQIARFVS GITFGLAVGG LNLRQQEQML KSRPDIVIAT 360
361 PGRFIDHIRN SASFNVDSVE ILVMDEADRM LEEGFQDELN EIMGLLPSNR QNLLFSATMN 420
421 SKIKSLVSLS LKKPVRIMID PPKKAATKLT QEFVRIRKRD HLKPALLFNL IRKLDPTGQK 480
481 RIVVFVARKE TAHRLRIIMG LLGMSVGELH GSLTQEQRLD SVNKFKNLEV PVLICTDLAS 540
541 RGLDIPKIEV VINYDMPKSY EIYLHRVGRT ARAGREGRSV TFVGESSQDR SIVRAAIKSV 600
601 EENKSLTQGK ALGRNVDWVQ IEETNKLVES MNDTIEDILV EEKEEKEILR AEMQLRKGEN 660
661 MLKHKKEIQA RPRRTWFQSE SDKKNSKVLG ALSRNKKVTN SKKRKREEAK ADGNGARSYR 720
721 KTKTDRIADQ ERTFKKQKST NSNKKKGFKS RR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
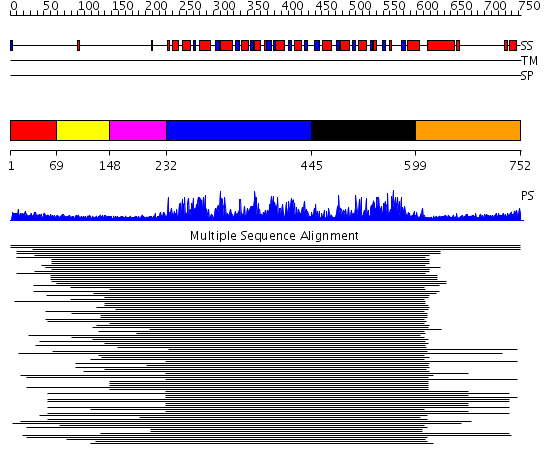
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 9.57 | No description for 1l8mA was found. | |
| 2 | View Details | [69..147] | 9.57 | No description for 1l8mA was found. | |
| 3 | View Details | [148..231] | 9.57 | No description for 1l8mA was found. | |
| 4 | View Details | [232..444] | 572.228787 | Putative DEAD box RNA helicase | |
| 5 | View Details | [445..598] | 572.228787 | Putative DEAD box RNA helicase | |
| 6 | View Details | [599..752] | 12.0 | Nucleotide excision repair enzyme UvrB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)