













| General Information: |
|
| Name(s) found: |
DPB2 /
YPR175W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 689 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[TAS nucleus [IDA cytoplasm [IDA epsilon DNA polymerase complex [IDA |
| Biological Process: |
lagging strand elongation
[TAS leading strand elongation [TAS nucleotide-excision repair [TAS mismatch repair [TAS |
| Molecular Function: |
DNA binding
[IEA]
nucleotidyltransferase activity [IEA] DNA-directed DNA polymerase activity [IEA][TAS] double-stranded DNA binding [IDA] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGSGNVLPV KIQPPLLRPL AYRVLSRKYG LSIKSDGLSA LAEFVGTNIG ANWRQGPATI 60
61 KFLEQFAAVW KQQERGLFID QSGVKEVIQE MKEREKVEWS HEHPIQHEEN ILGRTDDDEN 120
121 NSDDEMPIAA DSSLQNVSLS SPMRQPTERD EYKQPFKPES SKALDWRDYF KVINASQQQR 180
181 FSYNPHKMQF IFVPNKKQNG LGGIAGFLPD IEDKVQMFLT RYYLTNDRVM RNENFQNSDM 240
241 FNPLSSMVSL QNELSNTNRQ QQSSSMSITP IKNLLGRDAQ NFLLLGLLNK NFKGNWSLED 300
301 PSGSVEIDIS QTIPTQGHYY VPGCMVLVEG IYYSVGNKFH VTSMTLPPGE RREITLETIG 360
361 NLDLLGIHGI SNNNFIARLD KDLKIRLHLL EKELTDHKFV ILGANLFLDD LKIMTALSKI 420
421 LQKLNDDPPT LLIWQGSFTS VPVFASMSSR NISSSTQFKN NFDALATLLS RFDNLTENTT 480
481 MIFIPGPNDL WGSMVSLGAS GTLPQDPIPS AFTKKINKVC KNVVWSSNPT RIAYLSQEIV 540
541 IFRDDLSGRF KRHRLEFPFN ESEDVYTEND NMMSKDTDIV PIDELVKEPD QLPQKVQETR 600
601 KLVKTILDQG HLSPFLDSLR PISWDLDHTL TLCPIPSTMV LCDTTSAQFD LTYNGCKVIN 660
661 PGSFIHNRRA RYMEYVPSSK KTIQEEIYI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
DPB2 DPB3 DPB4 POL2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
CDC45 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 |
| View Details | Krogan NJ, et al. (2006) |
|
DPB2 DPB3 DPB4 INM1 MBA1 MCM2 POL2 RPN4 |
| View Details | Gavin AC, et al. (2006) |
|
DPB2 DPB4 POL2 |
| View Details | Gavin AC, et al. (2002) |
|
CTF18 CTF8 DPB2 DPB4 ELG1 ISW1 ISW2 KAP114 MOT1 NAP1 NPL6 OLA1 POL2 PUF6 RET1 RFC1 RFC2 RFC3 RFC4 RFC5 RPA135 RPC82 RPO31 STH1 TOP1 TOP2 VPS1 VPS13 YMR181C |
| View Details | Ho Y, et al. (2002) |

|
ATP3 AVO1 CCT5 CLU1 DHH1 DPB2 FUN12 GAL7 GPH1 IDH1 ILV2 MET16 MKK2 NOG2 NPA3 PDX1 PHO85 PUF3 RPN12 SIS1 SLX1 SRP54 STI1 TEM1 TFC7 VMA8 YER077C YGR266W YKR051W YKU80 YLR271W YPR003C |
| View Details | Qiu et al. (2008) |
|
CDC45 CDC9 DPB11 DPB2 DPB3 DPB4 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC2 ORC3 ORC5 POL1 POL12 POL2 POL3 POL30 POL31 POL32 PRI2 RAD30 RFC1 RFC2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Huh WK, et al. (2003) |
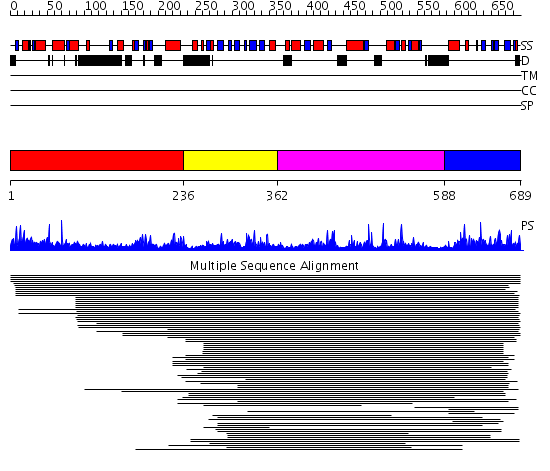
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [236..361] | 1.048988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [362..587] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [588..689] | 3.045983 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)