













| General Information: |
|
| Name(s) found: |
DBP7 /
YKR024C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 742 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear outer membrane
[IMP nucleolus [IDA |
| Biological Process: |
ribosome biogenesis
[RCA ribosomal large subunit assembly [IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDEDSMLLN FTTNEDTAGS SYKQAAKVTG GRWKDRRRMK MKLEGKTVSR KRKANTTGDE 60
61 GIIPGRGENS IKKLHKESSY SSEEQEKYKG RNAHNTQGRT LPADSQFVSS LFTSNREITT 120
121 AVNTNIHDEN VAINPSNAPL KGDQFASLGV SSLLVSHLEQ KMRIKKPTSI QKQAIPQIIG 180
181 NAGKNDFFIH AQTGSGKTLS YLLPIISTIL NMDTHVDRTS GAFALVIAPT RELASQIYHV 240
241 CSTLVSCCHY LVPCLLIGGE RKKSEKARLR KGCNFIIGTP GRVLDHLQNT KVIKEQLSQS 300
301 LRYIVLDEGD KLMELGFDET ISEIIKIVHD IPINSEKFPK LPHKLVHMLC SATLTDGVNR 360
361 LRNVALKDYK LISNGTKKDS DIVTVAPDQL LQRITIVPPK LRLVTLAATL NNITKDFIAS 420
421 GQQSKTLRTI VFVSCSDSVE FHYDAFSGSD GHHKNLTGDS VRLLTKGNTM FPCFSDSRDP 480
481 DVVIYKLHGS LSQQMRTSTL QHFARDNEAT KGKHLIMFCT DVASRGLDLP HVGSVIELDP 540
541 PFAVEDHLHR VGRTARAGEK GESLLFLLPG EEEKYMDYIQ PYHPMGWELL KFDKEILMPA 600
601 FKDVNVNRND KFIRKDEKSS KNKDVGDKEY EWDTNATTWH LNIERRVVGD SAFKNLAVKG 660
661 FISHVRAYAT HISQEKKFFN VKFLHLGHLA KSFGLRERPK AMGLQSSKDG NSEKKPTKEN 720
721 SKNKMFRMAR MAEKQIASEF NY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
DBP7 PUF6 PWP1 |
| View Details | Krogan NJ, et al. (2006) |

|
ALG5 BRE5 CSN12 CUP2 DBP7 LUG1 MET10 MSC7 MSE1 NOP53 NPY1 PBP4 RPF2 RRS1 SAS5 UME6 YGR126W YHR127W YPR045C YRA1 |
| View Details | Ho Y, et al. (2002) |

|
ARO1 BCK1 CBF5 CBP2 CCM1 CDC33 CHS1 CIC1 CMP2 DBP7 DRS1 EBP2 ENP2 ERB1 EST1 GBP2 HAS1 HMO1 HSH49 HTB1 HTB2 IMD1 IMD2 IMD3 ISA1 KAP95 KOG1 KRE33 KRI1 KRR1 KSP1 MGM101 MNP1 MRM1 MSS116 NOC2 NOP12 NOP13 NOP2 NOP58 NOP6 NOP7 NPL3 NUG1 PDI1 PET127 POL5 PRI2 PRP43 PSY2 PUF6 PWP1 RLI1 RPF2 RRP1 RRP12 RRP5 TIF1, TIF2 TIF4631 TIF6 TPD3 TRA1 TSR1 URB1 URB2 UTP10 UTP20 YER077C YPP1 YRA1 YTM1 |
| View Details | Qiu et al. (2008) |
|
AIR1 BFR2 BRX1 DBP10 DBP3 DBP6 DBP7 DBP8 DHR2 DRS1 ENP1 ERB1 FAL1 GAR1 HAS1 HCA4 IFH1 KRR1 MAK5 MRT4 MTR4 NAN1 NOC3 NOP1 NOP12 NOP13 NOP15 NOP4 NSR1 NUG1 POP1 PRP43 PUF6 PWP2 PXR1 RAT1 RCL1 RNT1 ROK1 RRP1 RRP15 RRP3 RRP8 SGD1 SMB1 SPB4 UTP10 UTP15 UTP18 UTP4 UTP5 UTP6 UTP8 UTP9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
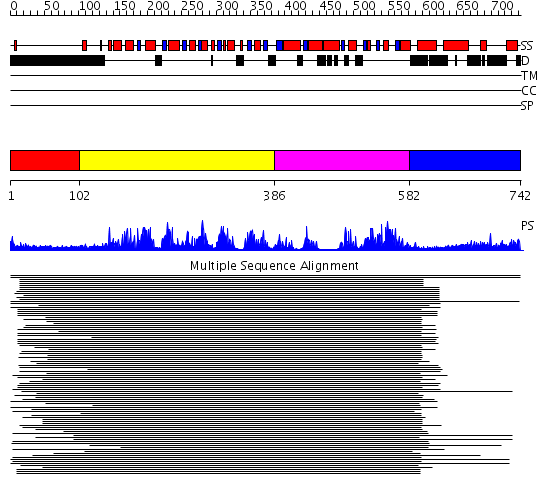
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [102..385] | 237.9897 | Putative DEAD box RNA helicase | |
| 3 | View Details | [386..581] | 237.9897 | Putative DEAD box RNA helicase | |
| 4 | View Details | [582..742] | 3.69897 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)