













| General Information: |
|
| Name(s) found: |
ILS1 /
YBL076C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1072 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
translation
[TAS |
| Molecular Function: |
isoleucine-tRNA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSESNAHFSF PKEEEKVLSL WDEIDAFHTS LELTKDKPEF SFFDGPPFAT GTPHYGHILA 60
61 STIKDIVPRY ATMTGHHVER RFGWDTHGVP IEHIIDKKLG ITGKDDVFKY GLENYNNECR 120
121 SIVMTYASDW RKTIGRLGRW IDFDNDYKTM YPSFMESTWW AFKQLHEKGQ VYRGFKVMPY 180
181 STGLTTPLSN FEAQQNYKDV NDPAVTIGFN VIGQEKTQLV AWTTTPWTLP SNLSLCVNAD 240
241 FEYVKIYDET RDRYFILLES LIKTLYKKPK NEKYKIVEKI KGSDLVGLKY EPLFPYFAEQ 300
301 FHETAFRVIS DDYVTSDSGT GIVHNAPAFG EEDNAACLKN GVISEDSVLP NAIDDLGRFT 360
361 KDVPDFEGVY VKDADKLIIK YLTNTGNLLL ASQIRHSYPF CWRSDTPLLY RSVPAWFVRV 420
421 KNIVPQMLDS VMKSHWVPNT IKEKRFANWI ANARDWNVSR NRYWGTPIPL WVSDDFEEVV 480
481 CVGSIKELEE LTGVRNITDL HRDVIDKLTI PSKQGKGDLK RIEEVFDCWF ESGSMPYASQ 540
541 HYPFENTEKF DERVPANFIS EGLDQTRGWF YTLAVLGTHL FGSVPYKNVI VSGIVLAADG 600
601 RKMSKSLKNY PDPSIVLNKY GADALRLYLI NSPVLKAESL KFKEEGVKEV VSKVLLPWWN 660
661 SFKFLDGQIA LLKKMSNIDF QYDDSVKSDN VMDRWILASM QSLVQFIHEE MGQYKLYTVV 720
721 PKLLNFIDEL TNWYIRFNRR RLKGENGVED CLKALNSLFD ALFTFVRAMA PFTPFLSESI 780
781 YLRLKEYIPE AVLAKYGKDG RSVHFLSYPV VKKEYFDEAI ETAVSRMQSV IDLGRNIREK 840
841 KTISLKTPLK TLVILHSDES YLKDVEALKN YIIEELNVRD VVITSDEAKY GVEYKAVADW 900
901 PVLGKKLKKD AKKVKDALPS VTSEQVREYL ESGKLEVAGI ELVKGDLNAI RGLPESAVQA 960
961 GQETRTDQDV LIIMDTNIYS ELKSEGLARE LVNRIQKLRK KCGLEATDDV LVEYELVKDT1020
1021 IDFEAIVKEH FDMLSKTCRS DIAKYDGSKT DPIGDEEQSI NDTIFKLKVF KL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
DED81 ILS1 |
| View Details | Krogan NJ, et al. (2006) |
|
FRS1 FRS2 ILS1 |
| View Details | Gavin AC, et al. (2006) |
|
DED81 GET3 ILS1 |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 GET3 ILS1 NIP1 UBP14 |
| View Details | Ho Y, et al. (2002) |

|
AAT2 ACC1 ACS2 ADE3 ADE6 ADH4 ADO1 AHA1 ALA1 APE2 ARA1 ARG4 ARO4 BAT1 BMS1 BUD13 CAR2 CDC10 CDC53 CDC60 CIC1 CLU1 CMD1 CMK2 CMP2 COF1 CPR1 CTF4 CYS3 DBP10 DED81 DIA2 DPS1 EDE1 ENP1 ERG10 ERG20 FPR3 GCY1 GPH1 GRS1 HCH1 HOM2 HYP2 ILS1 IMD3 IPP1 KRE33 KRI1 KRS1 LSP1 LST4 MCM2 MCM3 MCM5 MDH3 MPC54 MYO2 MYO4 MYO5 NAS6 NIP7 NMD3 NOP12 OLA1 PAB1 PDC5 PFK2 PGM2 PMI40 PML1 PRP6 PST2 RPA135 RPA190 RPC40 RRP12 SAC6 SCP160 SEH1 SHE3 SKP1 SLT2 SMC4 SOD1 SPB1 SRP1 SSF1 SSF2 SSZ1 SUI1 THR4 TIF6 TPS1 TRP5 TRR1 UBA1 URB1 URE2 UTP10 VAS1 VMA4 VPS13 YAK1 YBL104C YDL124W YNK1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
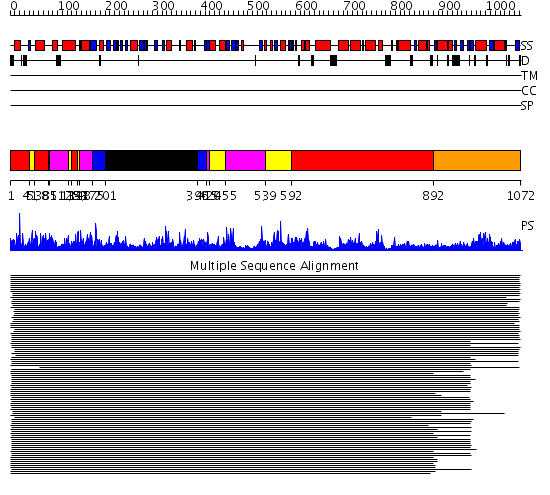
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..40] [53..80] [131..142] [592..891] |
2680.0 | Isoleucyl-tRNA synthetase (IleRS) | |
| 2 | View Details | [41..52] [81..84] [125..130] [143..147] [420..454] [539..591] |
2680.0 | Isoleucyl-tRNA synthetase (IleRS) | |
| 3 | View Details | [85..124] [148..174] [415..419] [455..538] |
2680.0 | Isoleucyl-tRNA synthetase (IleRS) | |
| 4 | View Details | [175..200] [396..414] |
2680.0 | Isoleucyl-tRNA synthetase (IleRS) | |
| 5 | View Details | [201..395] | 2680.0 | Isoleucyl-tRNA synthetase (IleRS) | |
| 6 | View Details | [892..1072] | 2.207964 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)