













| General Information: |
|
| Name(s) found: |
GET3 /
YDL100C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 354 amino acids |
Gene Ontology: |
|
| Cellular Component: |
GET complex
[IPI endoplasmic reticulum [IDA |
| Biological Process: |
response to heat
[IMP retrograde vesicle-mediated transport, Golgi to ER [IDA response to metal ion [IMP |
| Molecular Function: |
ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLTVEPNLH SLITSTTHKW IFVGGKGGVG KTTSSCSIAI QMALSQPNKQ FLLISTDPAH 60
61 NLSDAFGEKF GKDARKVTGM NNLSCMEIDP SAALKDMNDM AVSRANNNGS DGQGDDLGSL 120
121 LQGGALADLT GSIPGIDEAL SFMEVMKHIK RQEQGEGETF DTVIFDTAPT GHTLRFLQLP 180
181 NTLSKLLEKF GEITNKLGPM LNSFMGAGNV DISGKLNELK ANVETIRQQF TDPDLTTFVC 240
241 VCISEFLSLY ETERLIQELI SYDMDVNSII VNQLLFAEND QEHNCKRCQA RWKMQKKYLD 300
301 QIDELYEDFH VVKMPLCAGE IRGLNNLTKF SQFLNKEYNP ITDGKVIYEL EDKE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
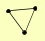
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
GET1 GET2 GET3 |
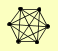
| View Details | Krogan NJ, et al. (2006) |
|
GET1 GET2 GET3 PSK1 TAT1 YHP1 |
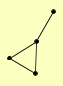
| View Details | Gavin AC, et al. (2006) |
|
CDC6 DED81 GET3 ILS1 |
| View Details | Gavin AC, et al. (2002) |

|
GET3 ILS1 |
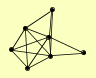
| View Details | Ho Y, et al. (2002) |
|
GET1 GET2 GET3 GRX7 GSF2 MSN4 POR1 |
| View Details | Qiu et al. (2008) |

|
GET1 GET3 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
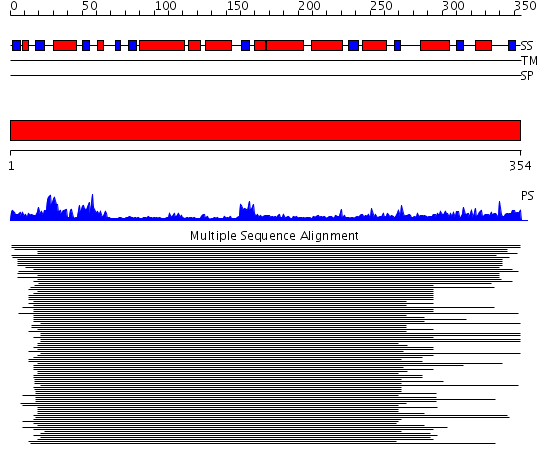
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 288.927757 | Arsenite-translocating ATPase ArsA |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)