













| General Information: |
|
| Name(s) found: |
SMC4 /
YLR086W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1418 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear condensin complex
[IPI |
| Biological Process: |
mitotic chromosome condensation
[IMP mitotic sister chromatid segregation [TAS |
| Molecular Function: |
ATPase activity
[ISS chromatin binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSPLSKRQ KRKSAQEPEL SLDQGDAEED SQVENRVNLS ENTPEPDLPA LEASYSKSYT 60
61 PRKLVLSSGE NRYAFSQPTN STTTSLHVPN LQPPKTSSRG RDHKSYSQSP PRSPGRSPTR 120
121 RLELLQLSPV KNSRVELQKI YDRHQSSSKQ QSRLFINELV LENFKSYAGK QVVGPFHTSF 180
181 SAVVGPNGSG KSNVIDSMLF VFGFRANKMR QDRLSDLIHK SEAFPSLQSC SVAVHFQYVI 240
241 DESSGTSRID EEKPGLIITR KAFKNNSSKY YINEKESSYT EVTKLLKNEG IDLDHKRFLI 300
301 LQGEVENIAQ MKPKAEKESD DGLLEYLEDI IGTANYKPLI EERMGQIENL NEVCLEKENR 360
361 FEIVDREKNS LESGKETALE FLEKEKQLTL LRSKLFQFKL LQSNSKLAST LEKISSSNKD 420
421 LEDEKMKFQE SLKKVDEIKA QRKEIKDRIS SCSSKEKTLV LERRELEGTR VSLEERTKNL 480
481 VSKMEKAEKT LKSTKHSISE AENMLEELRG QQTEHETEIK DLTQLLEKER SILDDIKLSL 540
541 KDKTKNISAE IIRHEKELEP WDLQLQEKES QIQLAESELS LLEETQAKLK KNVETLEEKI 600
601 LAKKTHKQEL QDLILDLKKK LNSLKDERSQ GEKNFTSAHL KLKEMQKVLN AHRQRAMEAR 660
661 SSLSKAQNKS KVLTALSRLQ KSGRINGFHG RLGDLGVIDD SFDVAISTAC PRLDDVVVDT 720
721 VECAQHCIDY LRKNKLGYAR FILLDRLRQF NLQPISTPEN VPRLFDLVKP KNPKFSNAFY 780
781 SVLRDTLVAQ NLKQANNVAY GKKRFRVVTV DGKLIDISGT MSGGGNHVAK GLMKLGTNQS 840
841 DKVDDYTPEE VDKIERELSE RENNFRVASD TVHEMEEELK KLRDHEPDLE SQISKAEMEA 900
901 DSLASELTLA EQQVKEAEMA YVKAVSDKAQ LNVVMKNLER LRGEYNDLQS ETKTKKEKIK 960
961 GLQDEIMKIG GIKLQMQNSK VESVCQKLDI LVAKLKKVKS ASKKSGGDVV KFQKLLQNSE1020
1021 RDVELSSDEL KVIEEQLKHT KLALAENDTN MNETLNLKVE LKEQSEQLKE QMEDMEESIN1080
1081 EFKSIEIEMK NKLEKLNSLL TYIKSEITQQ EKGLNELSIR DVTHTLGMLD DNKMDSVKED1140
1141 VKNNQELDQE YRSCETQDES EIKDAETSCD NYHPMNIDET SDEVSRGIPR LSEDELRELD1200
1201 VELIESKINE LSYYVEETNV DIGVLEEYAR RLAEFKRRKL DLNNAVQKRD EVKEQLGILK1260
1261 KKRFDEFMAG FNIISMTLKE MYQMITMGGN AELELVDSLD PFSEGVTFSV MPPKKSWRNI1320
1321 TNLSGGEKTL SSLALVFALH KYKPTPLYVM DEIDAALDFR NVSIVANYIK ERTKNAQFIV1380
1381 ISLRNNMFEL AQQLVGVYKR DNRTKSTTIK NIDILNRT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
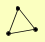
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
BRN1 SMC2 SMC4 |
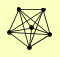
| View Details | Krogan NJ, et al. (2006) |
|
BRN1 HIS2 SMC2 SMC4 YCG1 YCS4 |
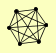
| View Details | Gavin AC, et al. (2002) |
|
DIS3 ISW1 MAS1 MAS2 RRP4 SMC4 |
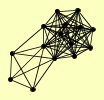
| View Details | Ho Y, et al. (2002) |
|
ACC1 AHA1 ARO4 BUD13 CDC60 ILS1 PML1 RPA135 RPA190 RPC40 SMC4 SRP1 TRP5 URE2 VMA4 |
| View Details | Qiu et al. (2008) |
|
ABF1 BRN1 HSK3 SMC4 TBF1 YCG1 YCS4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
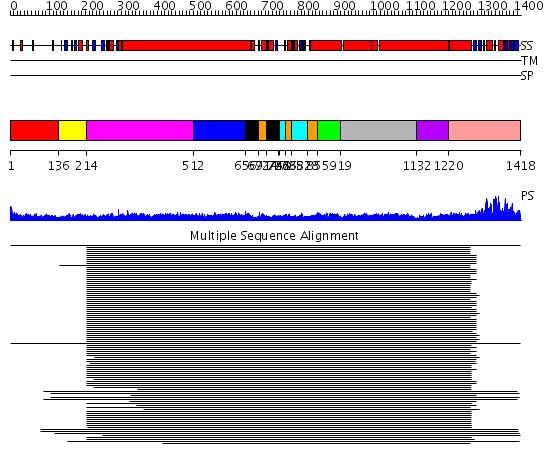
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [136..213] | 4.30103 | Rad50 | |
| 3 | View Details | [214..511] | 94.045757 | Heavy meromyosin subfragment | |
| 4 | View Details | [512..655] | 3.0 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 5 | View Details | [656..691] [714..746] |
9.69897 | Smc hinge domain | |
| 6 | View Details | [692..713] [747..750] [766..784] [828..854] |
9.69897 | Smc hinge domain | |
| 7 | View Details | [751..765] [785..827] |
9.69897 | Smc hinge domain | |
| 8 | View Details | [855..918] | 10.221849 | Tropomyosin | |
| 9 | View Details | [919..1131] | 10.221849 | Tropomyosin | |
| 10 | View Details | [1132..1219] | 3.522879 | Ribonuclease domain of colicin E3; Colicin E3 translocation domain; Colicin E3 receptor domain | |
| 11 | View Details | [1220..1418] | 170.751666 | Smc head domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)