













| General Information: |
|
| Name(s) found: |
LIA1 /
YJR070C
[SGD]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 325 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
ribosome biogenesis
[RCA microtubule cytoskeleton organization [IGI |
| Molecular Function: |
protein binding
[IPI deoxyhypusine monooxygenase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTNFEKHFQ ENVDECTLEQ LRDILVNKSG KTVLANRFRA LFNLKTVAEE FATKPEEAKK 60
61 AIEYIAESFV NDKSELLKHE VAYVLGQTKN LDAAPTLRHV MLDQNQEPMV RHEAAEALGA 120
121 LGDKDSLDDL NKAAKEDPHV AVRETCELAI NRINWTHGGA KDKENLQQSL YSSIDPAPPL 180
181 PLEKDATIPE LQALLNDPKQ PLFQRYRAMF RLRDIGTDEA ILALATGFSA ESSLFKHEIA 240
241 YVFGQIGSPA AVPSLIEVLG RKEEAPMVRH EAAEALGAIA SPEVVDVLKS YLNDEVDVVR 300
301 ESCIVALDMY DYENSNELEY APTAN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ARC35 ARG4 ARP2 AYR1 BGL2 CCT2 COR1 CPR6 ERG27 FAA4 GPT2 KGD2 LIA1 MAE1 OLA1 OM45 PCT1 PDA1 PET10 PET9 PIL1 PRE10 PRO3 RET2 RNR2 RPT4 RTN2 RVB2 RVS161 SEC18 SEC21 SEC26 SEC27 SEC28 SEC53 SEN2 SGD1 SHS1 TIF34 TIF35 TPA1 UTP5 YER138C YHR033W YNL181W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
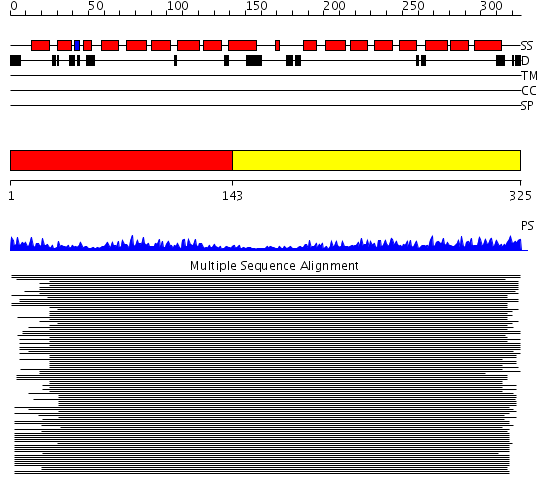
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | 40.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 2 | View Details | [143..325] | 40.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)