













| General Information: |
|
| Name(s) found: |
MGM1 /
YOR211C
[SGD]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 902 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial intermembrane space
[IDA mitochondrion [IDA |
| Biological Process: |
mitochondrial fusion
[IMP mitochondrion organization [IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
GTPase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSTSLRAI PRVANYNTLV RMNASPVRLL ILRRQLATHP AILYSSPYIK SPLVHLHSRM 60
61 SNVHRSAHAN ALSFVITRRS ISHFPKIISK IIRLPIYVGG GMAAAGSYIA YKMEEASSFT 120
121 KDKLDRIKDL GESMKEKFNK MFSGDKSQDG GHGNDGTVPT ATLIAATSLD DDESKRQGDP 180
181 KDDDDEDDDD EDDENDSVDT TQDEMLNLTK QMIEIRTILN KVDSSSAHLT LPSIVVIGSQ 240
241 SSGKSSVLES IVGREFLPKG SNMVTRRPIE LTLVNTPNSN NVTADFPSMR LYNIKDFKEV 300
301 KRMLMELNMA VPTSEAVSEE PIQLTIKSSR VPDLSLVDLP GYIQVEAADQ PIELKTKIRD 360
361 LCEKYLTAPN IILAISAADV DLANSSALKA SKAADPKGLR TIGVITKLDL VDPEKARSIL 420
421 NNKKYPLSMG YVGVITKTPS SINRKHLGLF GEAPSSSLSG IFSKGQHGQS SGEENTNGLK 480
481 QIVSHQFEKA YFKENKKYFT NCQVSTKKLR EKLIKILEIS MSNALEPTST LIQQELDDTS 540
541 YLFKVEFNDR HLTPKSYLLN NIDVLKLGIK EFQEKFHRNE LKSILRAELD QKVLDVLATR 600
601 YWKDDNLQDL SSSKLESDTD MLYWHKKLEL ASSGLTKMGI GRLSTMLTTN AILKELDNIL 660
661 ESTQLKNHEL IKDLVSNTAI NVLNSKYYST ADQVENCIKP FKYEIDLEER DWSLARQHSI 720
721 NLIKEELRQC NSRYQAIKNA VGSKKLANVM GYLENESNLQ KETLGMSKLL LERGSEAIFL 780
781 DKRCKVLSFR LKMLKNKCHS TIEKDRCPEV FLSAVSDKLT STAVLFLNVE LLSDFFYNFP 840
841 IELDRRLTLL GDEQVEMFAK EDPKISRHIE LQKRKELLEL ALEKIDSILV FKKSYKGVSK 900
901 NL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
FZO1 MGM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
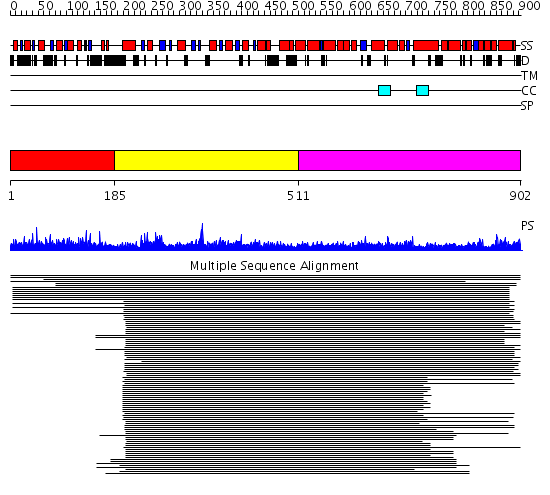
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 1.049939 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [185..510] | 506.529817 | Dynamin G domain | |
| 3 | View Details | [511..902] | 9.46 | Clathrin heavy chain proximal leg segment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|