













| General Information: |
|
| Name(s) found: |
YNR029C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 429 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSALRNIKFN EEEDGELPCL VTGEENNLQE ILENVSYDGG NIVSDAKVER VNKQVENTSA 60
61 GATDVHEKKR IPVSIITGYL GSGKSTLLEK IALKGADKKI AVILNEFGDS SEIEKAMTIK 120
121 NGSNSYQEWL DLGNGCLCCS LKNIGVKAIE DMVERSPGKI DYILLETSGI ADPAPIAKMF 180
181 WQDEGLNSSV YIDGIITVLD CEHILKCLDD ISIDAHWHGD KVGLEGNLTI AHFQLAMADR 240
241 IIMNKYDTIE HSPEMVKQLK ERVREINSIA PMFFTKYSDT PIQNLLDIHA YDSVRISDIL 300
301 DSGSGNGTIH DDRMGTIMLT FRPLKNEEEY NNKFIKQFLQ PLLWKNFGAM TVLGGRRRDD 360
361 GRDWEVQRTK GLILIEGENP IARVIQGVRD TYDVFPGKYD GSNKECKIVL IGKYLEKESI 420
421 EELLRKTLE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
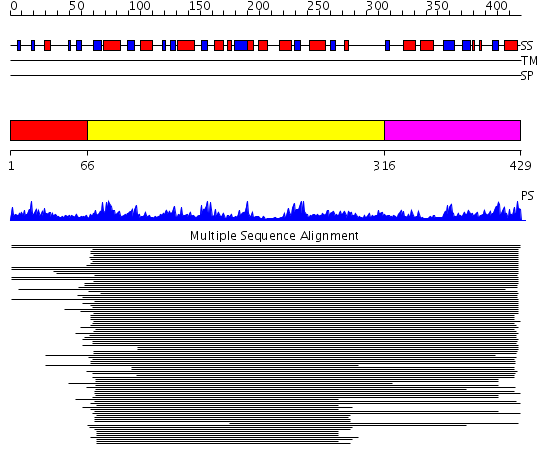
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [66..315] | 2.09691 | Signal sequence recognition protein Ffh; Signal sequence binding protein Ffh; GTPase domain of the signal sequence recognition protein Ffh | |
| 3 | View Details | [316..429] | 2.080988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)