













| General Information: |
|
| Name(s) found: |
PRP8 /
YHR165C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2413 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U4/U6 x U5 tri-snRNP complex
[IDA U5 snRNP [IDA |
| Biological Process: |
nuclear mRNA 3'-splice site recognition
[IMP nuclear mRNA splicing, via spliceosome [IPI |
| Molecular Function: |
U5 snRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGLPPPPPG FEEDSDLALP PPPPPPPGYE IEELDNPMVP SSVNEDTFLP PPPPPPSNFE 60
61 INAEEIVDFT LPPPPPPPGL DELETKAEKK VELHGKRKLD IGKDTFVTRK SRKRAKKMTK 120
121 KAKRSNLYTP KAEMPPEHLR KIINTHSDMA SKMYNTDKKA FLGALKYLPH AILKLLENMP 180
181 HPWEQAKEVK VLYHTSGAIT FVNETPRVIE PVYTAQWSAT WIAMRREKRD RTHFKRMRFP 240
241 PFDDDEPPLS YEQHIENIEP LDPINLPLDS QDDEYVKDWL YDSRPLEEDS KKVNGTSYKK 300
301 WSFDLPEMSN LYRLSTPLRD EVTDKNYYYL FDKKSFFNGK ALNNAIPGGP KFEPLYPREE 360
361 EEDYNEFNSI DRVIFRVPIR SEYKVAFPHL YNSRPRSVRI PWYNNPVSCI IQNDEEYDTP 420
421 ALFFDPSLNP IPHFIDNNSS LNVSNTKENG DFTLPEDFAP LLAEEEELIL PNTKDAMSLY 480
481 HSPFPFNRTK GKMVRAQDVA LAKKWFLQHP DEEYPVKVKV SYQKLLKNYV LNELHPTLPT 540
541 NHNKTKLLKS LKNTKYFQQT TIDWVEAGLQ LCRQGHNMLN LLIHRKGLTY LHLDYNFNLK 600
601 PTKTLTTKER KKSRLGNSFH LMRELLKMMK LIVDTHVQFR LGNVDAFQLA DGIHYILNHI 660
661 GQLTGIYRYK YKVMHQIRAC KDLKHIIYYK FNKNLGKGPG CGFWQPAWRV WLNFLRGTIP 720
721 LLERYIGNLI TRQFEGRSNE IVKTTTKQRL DAYYDLELRN SVMDDILEMM PESIRQKKAR 780
781 TILQHLSEAW RCWKANIPWD VPGMPAPIKK IIERYIKSKA DAWVSAAHYN RERIKRGAHV 840
841 EKTMVKKNLG RLTRLWIKNE QERQRQIQKN GPEITPEEAT TIFSVMVEWL ESRSFSPIPF 900
901 PPLTYKNDTK ILVLALEDLK DVYASKVRLN ASEREELALI EEAYDNPHDT LNRIKKYLLT 960
961 QRVFKPVDIT MMENYQNISP VYSVDPLEKI TDAYLDQYLW YEADQRKLFP NWIKPSDSEI1020
1021 PPLLVYKWTQ GINNLSEIWD VSRGQSAVLL ETTLGEMAEK IDFTLLNRLL RLIVDPNIAD1080
1081 YITAKNNVVI NFKDMSHVNK YGLIRGLKFA SFIFQYYGLV IDLLLLGQER ATDLAGPANN1140
1141 PNEFMQFKSK EVEKAHPIRL YTRYLDRIYM LFHFEEDEGE ELTDEYLAEN PDPNFENSIG1200
1201 YNNRKCWPKD SRMRLIRQDV NLGRAVFWEI QSRVPTSLTS IKWENAFVSV YSKNNPNLLF1260
1261 SMCGFEVRIL PRQRMEEVVS NDEGVWDLVD ERTKQRTAKA YLKVSEEEIK KFDSRIRGIL1320
1321 MASGSTTFTK VAAKWNTSLI SLFTYFREAI VATEPLLDIL VKGETRIQNR VKLGLNSKMP1380
1381 TRFPPAVFYT PKELGGLGMI SASHILIPAS DLSWSKQTDT GITHFRAGMT HEDEKLIPTI1440
1441 FRYITTWENE FLDSQRVWAE YATKRQEAIQ QNRRLAFEEL EGSWDRGIPR ISTLFQRDRH1500
1501 TLAYDRGHRI RREFKQYSLE RNSPFWWTNS HHDGKLWNLN AYRTDVIQAL GGIETILEHT1560
1561 LFKGTGFNSW EGLFWEKASG FEDSMQFKKL THAQRTGLSQ IPNRRFTLWW SPTINRANVY1620
1621 VGFLVQLDLT GIFLHGKIPT LKISLIQIFR AHLWQKIHES IVFDICQILD GELDVLQIES1680
1681 VTKETVHPRK SYKMNSSAAD ITMESVHEWE VSKPSLLHET NDSFKGLITN KMWFDVQLRY1740
1741 GDYDSHDISR YVRAKFLDYT TDNVSMYPSP TGVMIGIDLA YNMYDAYGNW FNGLKPLIQN1800
1801 SMRTIMKANP ALYVLRERIR KGLQIYQSSV QEPFLNSSNY AELFNNDIKL FVDDTNVYRV1860
1861 TVHKTFEGNV ATKAINGCIF TLNPKTGHLF LKIIHTSVWA GQKRLSQLAK WKTAEEVSAL1920
1921 VRSLPKEEQP KQIIVTRKAM LDPLEVHMLD FPNIAIRPTE LRLPFSAAMS IDKLSDVVMK1980
1981 ATEPQMVLFN IYDDWLDRIS SYTAFSRLTL LLRALKTNEE SAKMILLSDP TITIKSYHLW2040
2041 PSFTDEQWIT IESQMRDLIL TEYGRKYNVN ISALTQTEIK DIILGQNIKA PSVKRQKMAE2100
2101 LEAARSEKQN DEEAAGASTV MKTKTINAQG EEIVVVASAD YESQTFSSKN EWRKSAIANT2160
2161 LLYLRLKNIY VSADDFVEEQ NVYVLPKNLL KKFIEISDVK IQVAAFIYGM SAKDHPKVKE2220
2221 IKTVVLVPQL GHVGSVQISN IPDIGDLPDT EGLELLGWIH TQTEELKFMA ASEVATHSKL2280
2281 FADKKRDCID ISIFSTPGSV SLSAYNLTDE GYQWGEENKD IMNVLSEGFE PTFSTHAQLL2340
2341 LSDRITGNFI IPSGNVWNYT FMGTAFNQEG DYNFKYGIPL EFYNEMHRPV HFLQFSELAG2400
2401 DEELEAEQID VFS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
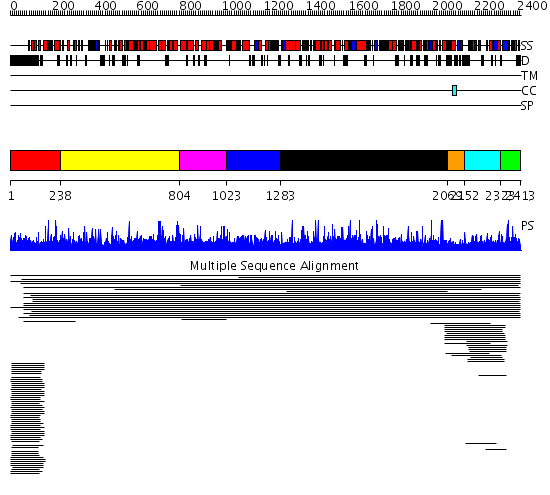
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..237] | 221.280999 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [238..803] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [804..1022] | 1.014889 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1023..1282] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1283..2068] | 1.022001 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [2069..2151] | N/A | Confident ab initio structure predictions are available. | |
| 7 | View Details | [2152..2322] | 15.053996 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [2323..2413] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1331..1527] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1528..1578] | 3.042987 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1579..1650] | 1.039962 | View MSA. No confident structure predictions are available. | |
| 12 | View Details | [1651..1743] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1744..1998] | 1.037955 | View MSA. No confident structure predictions are available. | |
| 14 | View Details | [1999..2070] | N/A | No confident structure predictions are available. | |
| 15 | View Details | [2071..2413] | 93.69897 | No description for 2og4A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)