













| General Information: |
|
| Name(s) found: |
MIS1 /
YBR084W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 975 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA folic acid biosynthetic process [IDA anaerobic purine catabolic process [IDA nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IMP acetate biosynthetic process from carbon monoxide [IDA |
| Molecular Function: |
formate-tetrahydrofolate ligase activity
[IDA methenyltetrahydrofolate cyclohydrolase activity [IDA methylenetetrahydrofolate dehydrogenase (NADP+) activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSRLSLLSN SRAFQQARWR IYRLKVSPTV HASQYHILSG RKLAQSIREK ANDEIQAIKL 60
61 KHPNFKPTLK IIQVGARPDS STYVRMKLKA SKDSNVDCII EKLPAEITEV ELLKKISDIN 120
121 DDDSIHGLLI QLPLPRHLDE TTITNAVDFK KDVDGFHRYN AGELAKKGGK PYFIPCTPYG 180
181 CMKLLEEAHV KLDGKNAVVL GRSSIVGNPI ASLLKNANAT VTVCHSHTRN IAEVVSQADI 240
241 VIAACGIPQY VKSDWIKEGA VVIDVGINYV PDISKKSGQK LVGDVDFDSV KEKTSYITPV 300
301 PGGVGPMTVA MLVSNVLLAA KRQFVESEKL PVIKPLPLHL ESPVPSDIDI SRAQSPKHIK 360
361 QVAEELGIHS HELELYGHYK AKISPNIFKR LESRENGKYV LVAGITPTPL GEGKSTTTMG 420
421 LVQALSAHLG KPSIANVRQP SLGPTLGVKG GAAGGGYAQV IPMDEFNLHL TGDIHAISAA 480
481 NNLLAAAIDT RMFHEATQKN DSTFYKRLVP RKKGIRKFTP SMQRRLKRLD IEKEDPDALT 540
541 PEEVKRFARL NINPDTITIR RVVDINDRML RQITIGEAAT EKGFTRTTGF DITVASELMA 600
601 ILALSKSLHE MKERIGRMVI GADYDNKPVT VEDIGCTGAL TALLRDAIKP NLMQTLEGTP 660
661 VMVHAGPFAN ISIGASSVIA DLMALKLVGS EKNPLNDKNI HEPGYVVTEA GFDFAMGGER 720
721 FFDIKCRSSG LVPDAVVLVA TVRALKSHGG APNVKPGQSL PKEYTEENID FVAKGVSNLV 780
781 KQIENIKTFG IPVVVAINRF ETDSQAEIEV IKKAALNAGA SHAVTSNHWM EGGKGAVELA 840
841 HAVVDATKEP KNFNFLYDVN SSIEDKLTSI VQKMYGGAKI EVSPEAQKKI DTYKKQGFGN 900
901 LPICIAKTQY SLSHDPSLKG VPRGFTFPIR DVRASIGAGY LYALAAEIQT IPGLSTYAGY 960
961 MAVEVDDDGE IEGLF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
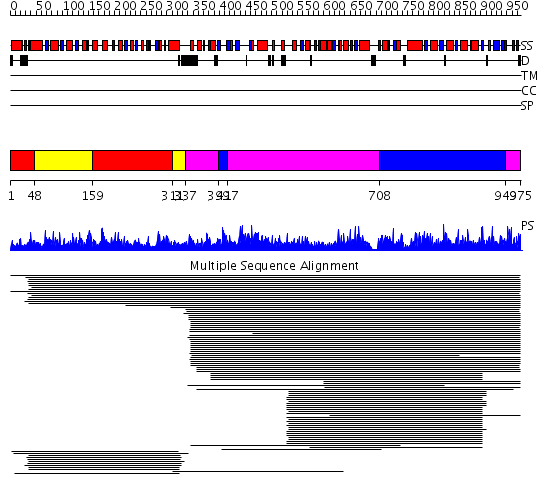
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..47] [159..310] |
844.0 | Methylenetetrahydrofolate dehydrogenase/cyclohydrolase; Tetrahydrofolate dehydrogenase/cyclohydrolase | |
| 2 | View Details | [48..158] [311..336] |
844.0 | Methylenetetrahydrofolate dehydrogenase/cyclohydrolase; Tetrahydrofolate dehydrogenase/cyclohydrolase | |
| 3 | View Details | [337..398] [417..707] [949..975] |
2420.0 | Formyltetrahydrofolate synthetase | |
| 4 | View Details | [399..416] [708..948] |
2420.0 | Formyltetrahydrofolate synthetase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)