













| General Information: |
|
| Name(s) found: |
FAA4 /
YMR246W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 694 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA cytoplasm [IDA |
| Biological Process: |
lipid transport
[IGI lipid metabolic process [IGI N-terminal protein myristoylation [IGI |
| Molecular Function: |
long-chain fatty acid-CoA ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEQYSVAVG EAANEHETAP RRNIRVKDQP LIRPINSSAS TLYEFALECF TKGGKRDGMA 60
61 WRDIIDIHET KKTIVKRVDG KDKPIEKTWL YYELTPYITM TYEEMICVMH DIGRGLIKIG 120
121 VKPNGENKFH IFASTSHKWM KTFLGCMSQG IPVVTAYDTL GESGLIHSMV ETDSVAIFTD 180
181 NQLLSKLAVP LKTAKNVKFV IHNEPIDPSD KRQNGKLYKA AKDAVDKIKE VRPDIKIYSF 240
241 DEIIEIGKKA KDEVELHFPK PEDPACIMYT SGSTGTPKGV VLTHYNIVAG IGGVGHNVIG 300
301 WIGPTDRIIA FLPLAHIFEL TFEFEAFYWN GILGYANVKT LTPTSTRNCQ GDLMEFKPTV 360
361 MVGVAAVWET VRKGILAKIN ELPGWSQTLF WTVYALKERN IPCSGLLSGL IFKRIREATG 420
421 GNLRFILNGG SAISIDAQKF LSNLLCPMLI GYGLTEGVAN ACVLEPEHFD YGIAGDLVGT 480
481 ITAKLVDVED LGYFAKNNQG ELLFKGAPIC SEYYKNPEET AAAFTDDGWF RTGDIAEWTP 540
541 KGQVKIIDRK KNLVKTLNGE YIALEKLESI YRSNPYVQNI CVYADENKVK PVGIVVPNLG 600
601 HLSKLAIELG IMVPGEDVES YIHEKKLQDA VCKDMLSTAK SQGLNGIELL CGIVFFEEEW 660
661 TPENGLVTSA QKLKRRDILA AVKPDVERVY KENT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2006) |
|
ACO1 FAA4 PGC1 |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ADR1 AFG2 AHA1 ALB1 ARP2 ARX1 BBC1 BFA1 BGL2 CCT6 CDC15 CDC33 CDC39 CDC53 CLU1 CPR6 CRM1 DSS4 DUG2 DUR1,2 EAP1 ECM29 ECM33 EXG1 FAA1 FAA4 FET3 GAL3 GCD1 GCD11 GCD2 GCD6 GCD7 GCN1 GCN3 GLT1 GSF2 HRT1 HYP2 LIA1 LOS1 MAE1 MDH2 MDN1 MET18 MKT1 MMS1 MYO2 NDE1 NMD5 NOG1 NOP7 NPA3 NUP53 OLA1 PDI1 PFK1 PMA2 PRO3 PSR2 RFC2 RMT2 RNR1 RNR2 RPA190 RPN1 RPN5 RPT4 SAC6 SAP155 SAP185 SAP190 SCW4 SEC4 SEC53 SGD1 SIT4 SNF4 SSD1 SUI3 TAP42 TEM1 TPS1 TRP2 UBI4 UTP5 VAC8 VMA8 VPS13 YAR009C YER138C YGP1 YIL108W YLR035C-A YMR196W YPT1 YTM1 |
| View Details | Qiu et al. (2008) |

|
FAA4 FAT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
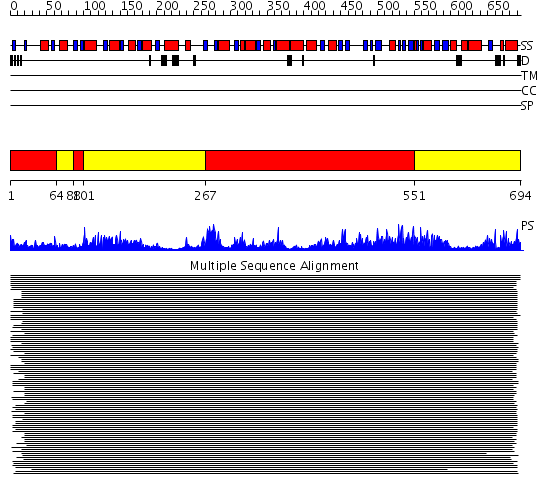
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..63] [88..100] [267..550] |
274.9794 | Luciferase | |
| 2 | View Details | [64..87] [101..266] [551..694] |
274.9794 | Luciferase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)