













| General Information: |
|
| Name(s) found: |
FBA1 /
YKL060C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 359 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
gluconeogenesis
[IMP glycolysis [IMP |
| Molecular Function: |
fructose-bisphosphate aldolase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVEQILKRK TGVIVGEDVH NLFTYAKEHK FAIPAINVTS SSTAVAALEA ARDSKSPIIL 60
61 QTSNGGAAYF AGKGISNEGQ NASIKGAIAA AHYIRSIAPA YGIPVVLHSD HCAKKLLPWF 120
121 DGMLEADEAY FKEHGEPLFS SHMLDLSEET DEENISTCVK YFKRMAAMDQ WLEMEIGITG 180
181 GEEDGVNNEN ADKEDLYTKP EQVYNVYKAL HPISPNFSIA AAFGNCHGLY AGDIALRPEI 240
241 LAEHQKYTRE QVGCKEEKPL FLVFHGGSGS TVQEFHTGID NGVVKVNLDT DCQYAYLTGI 300
301 RDYVLNKKDY IMSPVGNPEG PEKPNKKFFD PRVWVREGEK TMGAKITKSL ETFRTTNTL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ADH1 CAM1 EFT2, EFT1 ENO1 ENO2 FBA1 GPM1 HSC82 HSP104 HSP82 PDC1 PGK1 TDH2 TIF1, TIF2 TPI1 YEF3 |
| View Details | Gavin AC, et al. (2002) |

|
ADA2 ADH1 AMS1 ASC1 CDC14 COG3 CSE2 DHH1 ENO2 FAB1 FAR11 FAR3 FBA1 GAL11 GCN5 GPM1 HFM1 HSC82 MAF1 MED1 MED11 MED2 MED4 MED6 MED7 MED8 MLP1 MRPL6 NUT1 NUT2 PDC1 PGD1 PGK1 RET1 RGR1 ROM2 ROX3 RPA12 RPA135 RPA190 RPA43 RPA49 RPB10 RPB5 RPB8 RPC11 RPC17 RPC19 RPC25 RPC31 RPC34 RPC37 RPC40 RPC53 RPC82 RPG1 RPO21 RPO26 RPO31 RSC8 SIN4 SNT2 SPT7 SRB2 SRB4 SRB5 SRB6 SRB7 SRB8 SRO7 SRP1 SSE2 SSN2 SSN3 SSN8 TAF6 TIF1, TIF2 TPI1 YIL077C |
The following runs contain data for this protein:
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
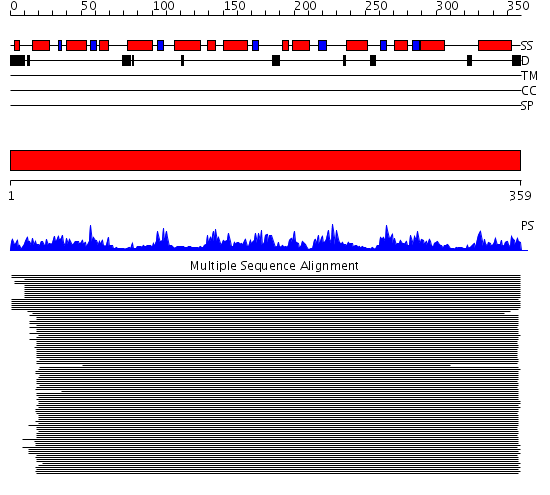
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..359] | 1114.0 | Fructose-bisphosphate aldolase (FBP aldolase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)