













| General Information: |
|
| Name(s) found: |
RPA43 /
YOR340C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 326 amino acids |
Gene Ontology: |
|
| Cellular Component: |
DNA-directed RNA polymerase I complex
[TAS |
| Biological Process: |
transcription from RNA polymerase I promoter
[TAS]
ribosome biogenesis [RCA |
| Molecular Function: |
DNA-directed RNA polymerase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQVKRANEN RETARFIKKH KKQVTNPIDE KNGTSNCIVR VPIALYVSLA PMYLENPLQG 60
61 VMKQHLNPLV MKYNNKVGGV VLGYEGLKIL DADPLSKEDT SEKLIKITPD TPFGFTWCHV 120
121 NLYVWQPQVG DVLEGYIFIQ SASHIGLLIH DAFNASIKKN NIPVDWTFVH NDVEEDADVI 180
181 NTDENNGNNN NEDNKDSNGG SNSLGKFSFG NRSLGHWVDS NGEPIDGKLR FTVRNVHTTG 240
241 RVVSVDGTLI SDADEEGNGY NSSRSQAESL PIVSNKKIVF DDEVSIENKE SHKELDLPEV 300
301 KEDNGSEIVY EENTSESNDG ESSDSD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
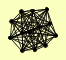
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
RPA12 RPA135 RPA14 RPA190 RPA34 RPA43 RPA49 RPB10 RPB5 RPB8 RPC19 RPC40 RPO26 |
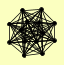
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RPA12 RPA135 RPA14 RPA190 RPA34 RPA43 RPA49 RPB10 RPB5 RPB8 RPC10 RPC19 RPC40 RPO26 |
| View Details | Krogan NJ, et al. (2006) |

|
BDP1 CDC14 COY1 FAR7 MAF1 RET1 RPA12 RPA135 RPA14 RPA190 RPA34 RPA43 RPA49 RPC10 RPC11 RPC17 RPC19 RPC25 RPC31 RPC34 RPC37 RPC40 RPC53 RPC82 RPO31 YDL144C YIR024C |
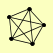
| View Details | Gavin AC, et al. (2006) |
|
RPA12 RPA190 RPA43 RPA49 RPC40 |
| View Details | Gavin AC, et al. (2002) |

|
AMS1 ASC1 CDC14 DHH1 FBA1 MAF1 RET1 RPA12 RPA135 RPA190 RPA43 RPA49 RPB10 RPB5 RPB8 RPC11 RPC17 RPC19 RPC25 RPC31 RPC34 RPC37 RPC40 RPC53 RPC82 RPO26 RPO31 RSC8 SRO7 |
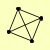
| View Details | Ho Y, et al. (2002) |
|
RPA135 RPA190 RPA43 RPB5 |
| View Details | Qiu et al. (2008) |

|
RET1 RPA12 RPA135 RPA14 RPA190 RPA34 RPA43 RPA49 RPB10 RPB11 RPB3 RPB5 RPB7 RPB8 RPC10 RPC19 RPC25 RPC31 RPC34 RPC37 RPC40 RPC82 RPO26 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
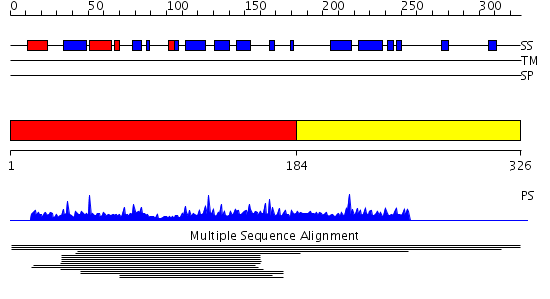
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 7.011978 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [184..326] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)