













| General Information: |
|
| Name(s) found: |
HAT1 /
YPL001W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 374 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA histone acetyltransferase complex [IDA |
| Biological Process: |
histone acetylation
[IDA chromatin silencing at telomere [IGI |
| Molecular Function: |
histone binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSANDFKPET WTSSANEALR VSIVGENAVQ FSPLFTYPIY GDSEKIYGYK DLIIHLAFDS 60
61 VTFKPYVNVK YSAKLGDDNI VDVEKKLLSF LPKDDVIVRD EAKWVDCFAE ERKTHNLSDV 120
121 FEKVSEYSLN GEEFVVYKSS LVDDFARRMH RRVQIFSLLF IEAANYIDET DPSWQIYWLL 180
181 NKKTKELIGF VTTYKYWHYL GAKSFDEDID KKFRAKISQF LIFPPYQNKG HGSCLYEAII 240
241 QSWLEDKSIT EITVEDPNEA FDDLRDRNDI QRLRKLGYDA VFQKHSDLSD EFLESSRKSL 300
301 KLEERQFNRL VEMLLLLNNS PSFELKVKNR LYIKNYDALD QTDPEKAREA LQNSFILVKD 360
361 DYRRIIESIN KSQG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
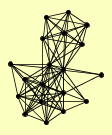
|
ASF1 HAT1 HAT2 HHF1, HHF2 HHT1, HHT2 HIF1 MAM33 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POB3 PSE1 PSH1 PTC4 RTT106 SPT16 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
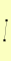
|
HAT1 HAT2 |
| View Details | Krogan NJ, et al. (2006) |
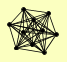
|
HAT1 HAT2 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 SWE1 YJR154W |
| View Details | Gavin AC, et al. (2006) |
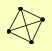
|
HAT1 HAT2 HHF1, HHF2 HIF1 |
| View Details | Gavin AC, et al. (2002) |
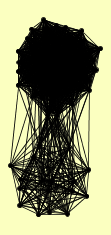
|
BDF2 CDC45 CDC55 CKB1 DYN1 ECM5 ERG26 GLT1 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 HSP42 ICL1 IML1 KAP123 KRE33 LIP2 LYS12 MAC1 MCM2 MCM5 MCM6 MCM7 MDN1 MIS1 MSB1 NIP1 NMD5 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPD3 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 SIN3 TEL1 TFP1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |
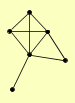
|
ELA1 GND1 HAT1 HAT2 HIF1 RTN1 |
| View Details | Qiu et al. (2008) |
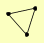
|
HAT1 SAS3 YNG1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
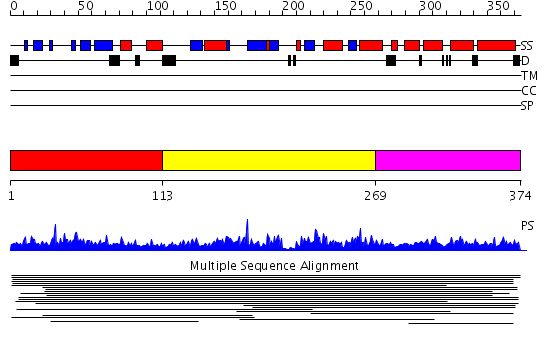
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 1865.0 | Histone acetyltransferase HAT1 | |
| 2 | View Details | [113..268] | 1865.0 | Histone acetyltransferase HAT1 | |
| 3 | View Details | [269..374] | 1865.0 | Histone acetyltransferase HAT1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)