













| General Information: |
|
| Name(s) found: |
YKU80 /
YMR106C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 629 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromatin
[TAS nuclear telomeric heterochromatin [TAS |
| Biological Process: |
double-strand break repair via homologous recombination
[IMP telomere maintenance [IMP chromatin assembly or disassembly [IDA chromatin silencing [IDA double-strand break repair via nonhomologous end joining [IDA donor selection [IDA |
| Molecular Function: |
DNA binding
[IDA RNA binding [IDA damaged DNA binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSESTTFIV DVSPSMMKNN NVSKSMAYLE YTLLNKSKKS RKTDWISCYL ANCPVSENSQ 60
61 EIPNVFQIQS FLAPVTTTAT IGFIKRLKQY CDQHSHDSSN EGLQSMIQCL LVVSLDIKQQ 120
121 FQARKILKQI VVFTDNLDDL DITDEEIDLL TEELSTRIIL IDCGKDTQEE RKKSNWLKLV 180
181 EAIPNSRIYN MNELLVEITS PATSVVKPVR VFSGELRLGA DILSTQTSNP SGSMQDENCL 240
241 CIKVEAFPAT KAVSGLNRKT AVEVEDSQKK ERYVGVKSII EYEIHNEGNK KNVSEDDQSG 300
301 SSYIPVTISK DSVTKAYRYG ADYVVLPSVL VDQTVYESFP GLDLRGFLNR EALPRYFLTS 360
361 ESSFITADTR LGCQSDLMAF SALVDVMLEN RKIAVARYVS KKDSEVNMCA LCPVLIEHSN 420
421 INSEKKFVKS LTLCRLPFAE DERVTDFPKL LDRTTTSGVP LKKETDGHQI DELMEQFVDS 480
481 MDTDELPEIP LGNYYQPIGE VTTDTTLPLP SLNKDQEENK KDPLRIPTVF VYRQQQVLLE 540
541 WIHQLMINDS REFEIPELPD SLKNKISPYT HKKFDSTKLV EVLGIKKVDK LKLDSELKTE 600
601 LEREKIPDLE TLLKRGEQHS RGSPNNSNN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
YKU70 YKU80 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
YKU70 YKU80 |
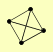
| View Details | Krogan NJ, et al. (2006) |
|
FMP48 MUC1 YKU70 YKU80 |
| View Details | Gavin AC, et al. (2006) |

|
YKU70 YKU80 |
| View Details | Gavin AC, et al. (2002) |

|
YKU70 YKU80 |
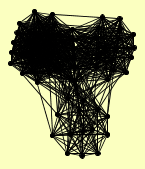
| View Details | Ho Y, et al. (2002) |
|
ACC1 ADR1 APT1 ARO1 ATP3 AVO1 CCT3 CCT5 CLU1 COP1 CPA2 DEF1 DHH1 DPB2 DSS1 ECM10 FOL2 FUN12 GAL7 GPH1 IDH1 ILV2 LSC1 LST8 LYS12 MET16 MKK2 MTC5 NOG2 NPA3 OYE2 PDX1 PHO85 PHO86 POR1 PRE1 PST2 PUF3 PUP3 RPN12 RRP3 SIP1 SIS1 SLC1 SLX1 SOD2 SRP54 STI1 TEM1 TFC7 TMA29 TPS1 TSA2 VMA8 YBT1 YER077C YGR266W YHR033W YKR051W YKU80 YLR271W YML020W YPR003C |
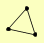
| View Details | Qiu et al. (2008) |
|
PCC1 TBF1 YKU80 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
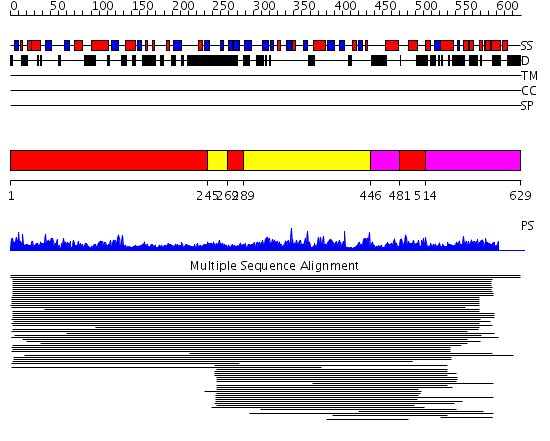
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..244] [269..288] [481..513] |
158.0 | DNA binding C-terminal domain of ku70; Ku70 subunit | |
| 2 | View Details | [245..268] [289..445] |
158.0 | DNA binding C-terminal domain of ku70; Ku70 subunit | |
| 3 | View Details | [446..480] [514..629] |
158.0 | DNA binding C-terminal domain of ku70; Ku70 subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)