













| General Information: |
|
| Name(s) found: |
ECO1 /
YFR027W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 281 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromatin
[IPI |
| Biological Process: |
DNA repair
[IDA DNA replication [IMP mitotic sister chromatid cohesion [IMP |
| Molecular Function: |
acetyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKARKSQRKA GSKPNLIQSK LQVNNGSKSN KIVKCDKCEM SYSSTSIEDR AIHEKYHTLQ 60
61 LHGRKWSPNW GSIVYTERNH SRTVHLSRST GTITPLNSSP LKKSSPSITH QEEKIVYVRP 120
121 DKSNGEVRAM TEIMTLVNNE LNAPHDENVI WNSTTEEKGK AFVYIRNDRA VGIIIIENLY 180
181 GGNGKTSSRG RWMVYDSRRL VQNVYPDFKI GISRIWVCRT ARKLGIATKL IDVARENIVY 240
241 GEVIPRYQVA WSQPTDSGGK LASKYNGIMH KSGKLLLPVY I |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
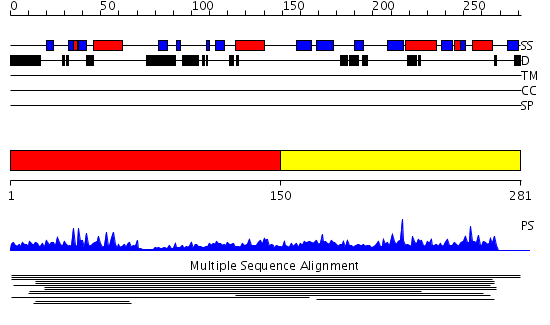
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 2.010983 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [150..281] | 1.009969 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)