













| General Information: |
|
| Name(s) found: |
CRM1 /
YGR218W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1084 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
mRNA export from nucleus
[IMP ribosomal large subunit export from nucleus [IMP protein export from nucleus [IMP |
| Molecular Function: |
protein transmembrane transporter activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGILDFSND LDIALLDQVV STFYQGSGVQ QKQAQEILTK FQDNPDAWQK ADQILQFSTN 60
61 PQSKFIALSI LDKLITRKWK LLPNDHRIGI RNFVVGMIIS MCQDDEVFKT QKNLINKSDL 120
121 TLVQILKQEW PQNWPEFIPE LIGSSSSSVN VCENNMIVLK LLSEEVFDFS AEQMTQAKAL 180
181 HLKNSMSKEF EQIFKLCFQV LEQGSSSSLI VATLESLLRY LHWIPYRYIY ETNILELLST 240
241 KFMTSPDTRA ITLKCLTEVS NLKIPQDNDL IKRQTVLFFQ NTLQQIATSV MPVTADLKAT 300
301 YANANGNDQS FLQDLAMFLT TYLARNRALL ESDESLRELL LNAHQYLIQL SKIEERELFK 360
361 TTLDYWHNLV ADLFYEVQRL PATEMSPLIQ LSVGSQAIST GSGALNPEYM KRFPLKKHIY 420
421 EEICSQLRLV IIENMVRPEE VLVVENDEGE IVREFVKESD TIQLYKSERE VLVYLTHLNV 480
481 IDTEEIMISK LARQIDGSEW SWHNINTLSW AIGSISGTMS EDTEKRFVVT VIKDLLDLTV 540
541 KKRGKDNKAV VASDIMYVVG QYPRFLKAHW NFLRTVILKL FEFMHETHEG VQDMACDTFI 600
601 KIVQKCKYHF VIQQPRESEP FIQTIIRDIQ KTTADLQPQQ VHTFYKACGI IISEERSVAE 660
661 RNRLLSDLMQ LPNMAWDTIV EQSTANPTLL LDSETVKIIA NIIKTNVAVC TSMGADFYPQ 720
721 LGHIYYNMLQ LYRAVSSMIS AQVAAEGLIA TKTPKVRGLR TIKKEILKLV ETYISKARNL 780
781 DDVVKVLVEP LLNAVLEDYM NNVPDARDAE VLNCMTTVVE KVGHMIPQGV ILILQSVFEC 840
841 TLDMINKDFT EYPEHRVEFY KLLKVINEKS FAAFLELPPA AFKLFVDAIC WAFKHNNRDV 900
901 EVNGLQIALD LVKNIERMGN VPFANEFHKN YFFIFVSETF FVLTDSDHKS GFSKQALLLM 960
961 KLISLVYDNK ISVPLYQEAE VPQGTSNQVY LSQYLANMLS NAFPHLTSEQ IASFLSALTK1020
1021 QYKDLVVFKG TLRDFLVQIK EVGGDPTDYL FAEDKENALM EQNRLEREKA AKIGGLLKPS1080
1081 ELDD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
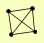
|
CRM1 GEA2 MIR1 YHR020W |
| View Details | Gavin AC, et al. (2002) |
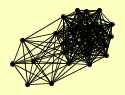
|
BMH1 BMH2 CLU1 CRM1 ECM29 FKS1 GCN1 GEA2 GFA1 HRP1 HSP42 KAP104 KAP123 LCB1 LCB2 LYS12 MIR1 NAB2 NSL1 PDC1 PRP11 SAM1 SAM2 TAT1 YHR020W |
| View Details | Ho Y, et al. (2002) |

|
AAC3 ADR1 ATG12 BBC1 BUD32 CAR2 CDC39 CDC53 CPR6 CRM1 DIA4 DUR1,2 ECM29 ECM33 FAA4 FET3 GAL3 GCN1 GRX3 GRX4 HEF3 HRT1 HYP2 IDP2 IMD2 IMD4 KAE1 KAP122 KGD1 MDN1 MET10 MET18 MKT1 MMS1 MYO2 NPA3 PFK1 PHO81 PMA2 RNH202 RPA190 RPN1 RPN5 RPN6 RPT1 SEC18 SEC23 SGO1 TMA29 TPS1 UBI4 URA7 VPS13 YAR009C YGP1 YHR033W YLR035C-A YRB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
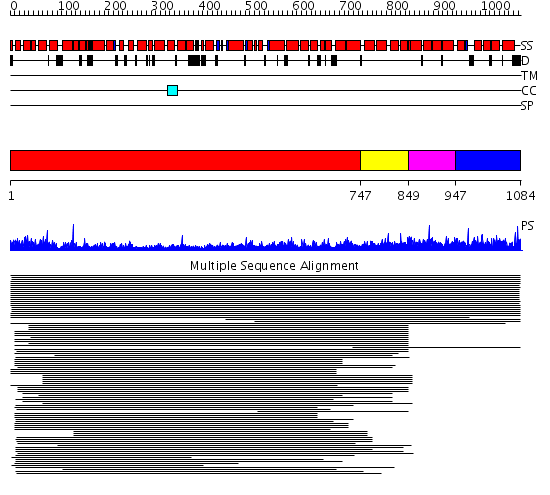
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..746] | 33.191997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [747..848] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [849..946] | 8.41 | Hemoglobin, alpha-chain | |
| 4 | View Details | [947..1084] | 8.41 | Hemoglobin, alpha-chain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)