













| General Information: |
|
| Name(s) found: |
SAC6 /
YDR129C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
actin cortical patch
[IDA |
| Biological Process: |
bipolar cellular bud site selection
[IMP response to osmotic stress [IMP actin filament organization [IMP endocytosis [IMP |
| Molecular Function: |
actin filament binding
[IDA protein binding, bridging [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNIVKLQRKF PILTQEDLFS TIEKFRAIDL DDKGWVEKQQ ALEAVSKDGD ATYDEARETL 60
61 KHVGVDASGR VELDDYVGLV AKLRESKTGA APQTTFNVAP NSTPIVSTAA TGLQHKGKGT 120
121 QAKIIVAGSQ TGTTHTINEE ERREFTKHIN SVLAGDQDIG DLLPFPTDTF QLFDECRDGL 180
181 VLSKLINDSV PDTIDTRVLN WPKKGKELNN FQASENANIV INSAKAIGCV VVNVHSEDII 240
241 EGREHLILGL IWQIIRRGLL SKIDIKLHPE LYRLLEDDET LEQFLRLPPE QILLRWFNYH 300
301 LKQANWNRRV TNFSKDVSDG ENYTILLNQL DPALCSKAPL QTTDLMERAE QVLQNAEKLD 360
361 CRKYLTPSSL VAGNPKLNLA FVAHLFNTHP GLEPIQEEEK PEIEEFDAEG EREARVFTLW 420
421 LNSLDVDPPV ISLFDDLKDG LILLQAYEKV MPGAVDFKHV NKRPASGAEI SRFKALENTN 480
481 YAVDLGRAKG FSLVGIEGSD IVDGNKLLTL GLVWQLMRRN ISITMKTLSS SGRDMSDSQI 540
541 LKWAQDQVTK GGKNSTIRSF KDQALSNAHF LLDVLNGIAP GYVDYDLVTP GNTEEERYAN 600
601 ARLAISIARK LGALIWLVPE DINEVRARLI ITFIASLMTL NK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ABP1 CMD1 CRN1 ECM10 EFT2, EFT1 LSP1 MLC1 RVS161 SAC6 TWF1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
ABP1 AIP1 ARC40 ARP2 BEM1 BUD6 CAP1 CAP2 COF1 CRN1 DLD2 GLK1 LAS17 OYE2 PAN1 PFY1 RVS167 SAC6 SLA1 SLA2 SRV2 TPM1 TPM2 TWF1 VRP1 |
| View Details | Krogan NJ, et al. (2006) |
|
ARC15 ASR1 CAJ1 CDC7 CRN1 INP52 IRC22 MSK1 PNO1 PST2 RAD34 SAC6 SAR1 SLM1 TCP1 TED1 YPP1 |
| View Details | Gavin AC, et al. (2006) |
|
ACT1 SAC6 |
| View Details | Ho Y, et al. (2002) |

|
ACS2 ADE3 ADE6 ADO1 ALA1 APE2 ARA1 BAT1 CDC10 CLU1 CYS3 DED81 DPS1 ERG10 ERG20 FAA4 FPR1 GCY1 GPH1 GRS1 HOM2 HYP2 ILS1 IMD3 KRS1 LSP1 MDH1 MDH3 OLA1 PAB1 PFK2 PGM2 PMI40 PRP6 PSR2 SAC6 SCP160 SOD1 SSZ1 THR4 TRP5 TRR1 UBA1 YDL124W |
| View Details | Qiu et al. (2008) |
|
ABP1 ACT1 AIP1 ARC40 ARP2 BBC1 BSP1 BUD6 CAP1 CAP2 COF1 CRN1 EDE1 END3 ENT1 HOF1 LAS17 MYO2 MYO5 PAN1 PFY1 RVS161 RVS167 SAC6 SCP1 SLA1 SLA2 SRV2 TPM1 TPM2 TWF1 VRP1 YSC84 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
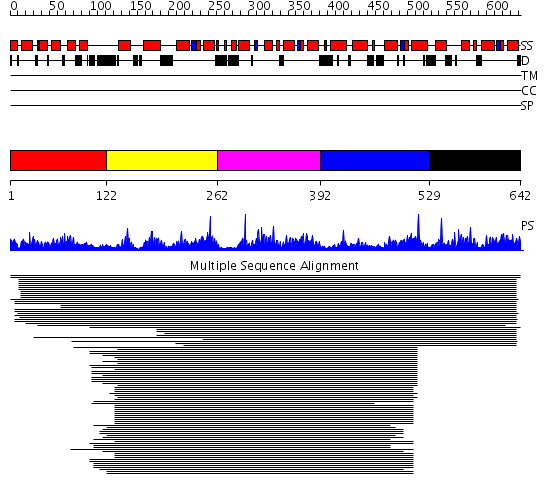
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 45.045757 | Calmodulin | |
| 2 | View Details | [122..261] | 693.450636 | N-terminal actin-crosslinking domain from fimbrin | |
| 3 | View Details | [262..391] | 693.450636 | N-terminal actin-crosslinking domain from fimbrin | |
| 4 | View Details | [392..528] | 6.045757 | Genetically engineered molecular motor | |
| 5 | View Details | [529..642] | 14.99 | beta-spectrin |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)