













| General Information: |
|
| Name(s) found: |
FAP7 /
YDL166C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 197 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IMP [NOT]preribosome [IPI |
| Biological Process: |
[NOT]ribosomal small subunit export from nucleus
[IMP response to oxidative stress [IMP maturation of SSU-rRNA [IMP |
| Molecular Function: |
nucleoside-triphosphatase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEARRYGPNI IVTGTPGCGK SSTCEFLKNK LKDYKYYNIS DFAKDNDCFE GYDEGRKSHI 60
61 VDEDKLLDML EPLLRQGNSI VDWHVNDVFP ERLIDLVVVL RCDNSNLYSR LHARGYHDSK 120
121 IEENLDAEIM GVVKQDAVES YEPHIVVELQ SDTKEDMVSN VSRIVAWEKM WLEQHPDGVT 180
181 NEYQGPRSDD EDDEDSE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
ADE4 ASC1 BFR2 BMS1 BRX1 BUD21 CBF5 CKA1 CKA2 CKB1 CKB2 CMS1 DBP8 DIA4 DIM1 DIP2 DIS3 ECM16 ECM29 EMG1 ENP1 ENP2 ERO1 ESF1 ESF2 FAP7 GCN1 HCA4 HHF1, HHF2 HSL1 HTA2 IMP3 IMP4 KRE33 KRR1 LCP5 LTV1 MPP10 MRD1 MVD1 NAN1 NOB1 NOC2 NOC4 NOP1 NOP14 NOP56 NOP58 NOP6 PNO1 PRE6 PRE9 PRP43 PWP2 RCL1 RML2 ROK1 RPP2B RRP12 RRP7 RRP9 RTG2 SAM1 SCL1 SEC21 SLX9 SOF1 TIF1, TIF2 TSR1 UTP10 UTP11 UTP13 UTP15 UTP18 UTP20 UTP21 UTP22 UTP30 UTP4 UTP6 UTP7 UTP8 UTP9 YOR059C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
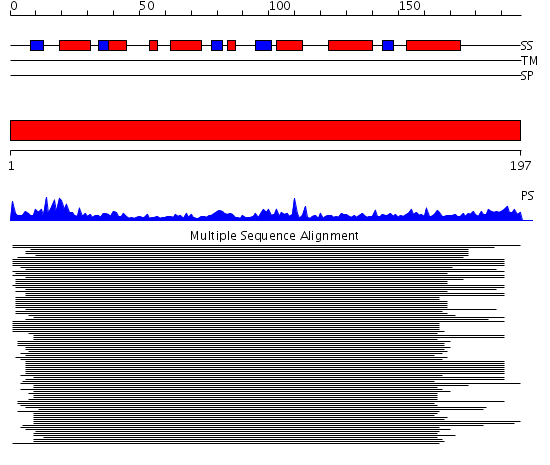
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 28.0 | Adenylate kinase; Microbial and mitochondrial ADK, insert "zinc finger" domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)