













| General Information: |
|
| Name(s) found: |
SSA1 /
YAL005C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 642 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA chaperonin-containing T-complex [IPI fungal-type cell wall [IDA cytoplasm [IDA fungal-type vacuole membrane [IDA |
| Biological Process: |
protein targeting to mitochondrion
[IMP SRP-dependent cotranslational protein targeting to membrane, translocation [IDA protein import into nucleus, translocation [IDA protein folding [IDA protein refolding [IDA response to stress [IMP translation [IMP |
| Molecular Function: |
ATPase activity
[IDA unfolded protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKAVGIDLG TTYSCVAHFA NDRVDIIAND QGNRTTPSFV AFTDTERLIG DAAKNQAAMN 60
61 PSNTVFDAKR LIGRNFNDPE VQADMKHFPF KLIDVDGKPQ IQVEFKGETK NFTPEQISSM 120
121 VLGKMKETAE SYLGAKVNDA VVTVPAYFND SQRQATKDAG TIAGLNVLRI INEPTAAAIA 180
181 YGLDKKGKEE HVLIFDLGGG TFDVSLLFIE DGIFEVKATA GDTHLGGEDF DNRLVNHFIQ 240
241 EFKRKNKKDL STNQRALRRL RTACERAKRT LSSSAQTSVE IDSLFEGIDF YTSITRARFE 300
301 ELCADLFRST LDPVEKVLRD AKLDKSQVDE IVLVGGSTRI PKVQKLVTDY FNGKEPNRSI 360
361 NPDEAVAYGA AVQAAILTGD ESSKTQDLLL LDVAPLSLGI ETAGGVMTKL IPRNSTISTK 420
421 KFEIFSTYAD NQPGVLIQVF EGERAKTKDN NLLGKFELSG IPPAPRGVPQ IEVTFDVDSN 480
481 GILNVSAVEK GTGKSNKITI TNDKGRLSKE DIEKMVAEAE KFKEEDEKES QRIASKNQLE 540
541 SIAYSLKNTI SEAGDKLEQA DKDTVTKKAE ETISWLDSNT TASKEEFDDK LKELQDIANP 600
601 IMSKLYQAGG APGGAAGGAP GGFPGGAPPA PEAEGPTVEE VD |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
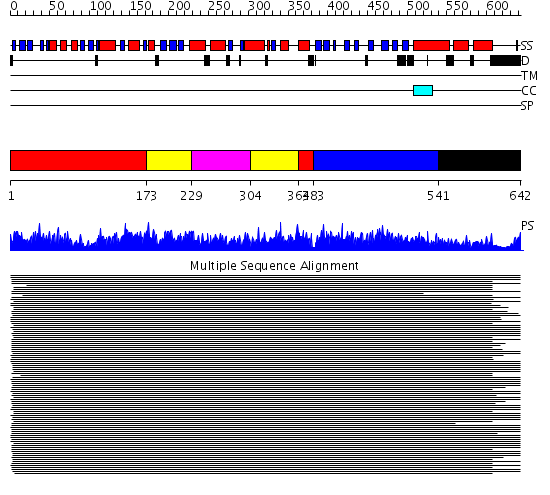
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] [364..382] |
2770.0 | Heat shock protein 70kDa, ATPase fragment | |
| 2 | View Details | [173..228] [304..363] |
2770.0 | Heat shock protein 70kDa, ATPase fragment | |
| 3 | View Details | [229..303] | 2770.0 | Heat shock protein 70kDa, ATPase fragment | |
| 4 | View Details | [383..540] | 734.0 | DnaK | |
| 5 | View Details | [541..642] | 8.94 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)