













| General Information: |
|
| Name(s) found: |
CTR9 /
YOL145C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1077 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA transcription elongation factor complex [IPI Cdc73/Paf1 complex [IPI |
| Biological Process: |
histone methylation
[IMP transcription from RNA polymerase II promoter [IPI chromosome segregation [IMP RNA elongation from RNA polymerase II promoter [IPI |
| Molecular Function: |
RNA polymerase II transcription elongation factor activity
[IPI triplex DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNAMKVEGY PSMEWPTSLD IPLKASEELV GIDLETDLPD DPTDLKTLLV EENSEKEHWL 60
61 TIALAYCNHG KTNEGIKLIE MALDVFQNSE RASLHTFLTW AHLNLAKGQS LSVETKEHEL 120
121 TQAELNLKDA IGFDPTWIGN MLATVELYYQ RGHYDKALET SDLFVKSIHA EDHRSGRQSK 180
181 PNCLFLLLRA KLLYQKENYM ASLKIFQELL VINPVLQPDP RIGIGLCFWQ LKDSKMAIKS 240
241 WQRALQLNPK NTSASILVLL GEFRESFTNS TNDKTFKEAF TKALSDLNNI FSENQHNPVL 300
301 LTLLQTYYYF KGDYQTVLDI YHHRILKMSP MIAKIVLSES SFWCGRAHYA LGDYRKSFIM 360
361 FQESLKKNED NLLAKLGLGQ TQIKNNLLEE SIITFENLYK TNESLQELNY ILGMLYAGKA 420
421 FDAKTAKNTS AKEQSNLNEK ALKYLERYLK LTLATKNQLV ISRAYLVISQ LYELQNQYKT 480
481 SLDYLSKALE EMEFIKKEIP LEVLNNLACY HFINGDFIKA DDLFKQAKAK VSDKDESVNI 540
541 TLEYNIARTN EKNDCEKSES IYSQVTSLHP AYIAARIRNL YLKFAQSKIE DSDMSTEMNK 600
601 LLDLNKSDLE IRSFYGWYLK NSKERKNNEK STTHNKETLV KYNSHDAYAL ISLANLYVTI 660
661 ARDGKKSRNP KEQGKSKHSY LKAIQLYQKV LQVDPFNIFA AQGLAIIFAE SKRLGPALEI 720
721 LRKVRDSLDN EDVQLNLAHC YLEMREYGKA IENYELVLKK FDNEKTRPHI LNLLGRAWYA 780
781 RAIKETSVNF YQKALENAKT ALDLFVKESS KSKFIHSVKF NIALLHFQIA ETLRRSNPKF 840
841 RTVQQIKDSL EGLKEGLELF RELNDLKEFN MIPKEELEQR IQLGETTMKS ALERSLNEQE 900
901 EFKKEQRAKI DEARKILEEN ELKEQGWMKQ EEEARRLKLE KQAEEYRKLQ DEAQKIFQVR 960
961 EAMAISEHNV KDDSDLSDKD NEYDEEQPRQ KRKRSTKTKN SGESKRRKAA KKTLSDSDED1020
1021 DDDVVKKPSH NKGKKSQLSN EFIEDSDEEE AQMSGSEQNK NDDNDENNDN DDNDGLF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC73 CKA1 CKA2 CKB1 CKB2 CTR9 LEO1 PAF1 POB3 RTF1 SPT16 |
| View Details | Krogan NJ, et al. (2006) |
|
ARA2 AYT1 BRO1 CDC73 CTR9 GLO4 HHT1, HHT2 LEO1 PAF1 PML39 POB3 PSH1 RTF1 SEN1 SPT16 YHR035W |
| View Details | Gavin AC, et al. (2002) |

|
AVO1 BEM2 CDC73 CEG1 CET1 CKA1 CKA2 CKB1 CKB2 CLU1 CMD1 CSL4 CTR9 DIS3 ECM16 GCN1 HHF1, HHF2 IML1 LEO1 LSM12 LYS12 MAM33 MLC1 MTR3 MYO2 NIP1 NMD5 NOP1 PAF1 PDB1 POB3 PRP43 RPG1 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 RTF1 SAM1 SHE4 SKI6 SKI7 SPT16 SRP1 SSA3 SSA4 TOP2 UTP10 VPS1 |
| View Details | Qiu et al. (2008) |
|
CDC73 CTR9 ELC1 LEO1 PAF1 SPN1 SPT4 SPT6 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
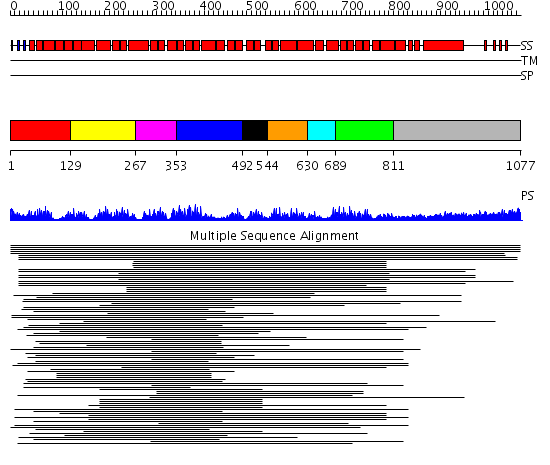
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 2.39794 | Hop | |
| 2 | View Details | [129..266] | 33.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [267..352] | 33.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [353..491] | 33.69897 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [492..543] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [544..629] | 2.30103 | Protein farnesyltransferase alpha-subunit | |
| 7 | View Details | [630..688] | 2.30103 | Protein farnesyltransferase alpha-subunit | |
| 8 | View Details | [689..810] | 4.221849 | Heavy meromyosin subfragment | |
| 9 | View Details | [811..1077] | 4.221849 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)