













| General Information: |
|
| Name(s) found: |
TOP1 /
YOL006C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 769 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IPI |
| Biological Process: |
mitotic chromosome condensation
[IGI chromatin silencing at rDNA [IMP DNA strand elongation involved in DNA replication [IMP nuclear migration [IGI regulation of mitotic recombination [IMP chromatin assembly or disassembly [IMP DNA topological change [IDA regulation of transcription from RNA polymerase II promoter [IMP RNA elongation from RNA polymerase II promoter [IMP |
| Molecular Function: |
DNA topoisomerase type I activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTIADASKVN HELSSDDDDD VPLSQTLKKR KVASMNSASL QDEAEPYDSD EAISKISKKK 60
61 TKKIKTEPVQ SSSLPSPPAK KSATSKPKKI KKEDGDVKVK TTKKEEQENE KKKREEEEEE 120
121 DKKAKEEEEE YKWWEKENED DTIKWVTLKH NGVIFPPPYQ PLPSHIKLYY DGKPVDLPPQ 180
181 AEEVAGFFAA LLESDHAKNP VFQKNFFNDF LQVLKESGGP LNGIEIKEFS RCDFTKMFDY 240
241 FQLQKEQKKQ LTSQEKKQIR LEREKFEEDY KFCELDGRRE QVGNFKVEPP DLFRGRGAHP 300
301 KTGKLKRRVN PEDIVLNLSK DAPVPPAPEG HKWGEIRHDN TVQWLAMWRE NIFNSFKYVR 360
361 LAANSSLKGQ SDYKKFEKAR QLKSYIDAIR RDYTRNLKSK VMLERQKAVA IYLIDVFALR 420
421 AGGEKSEDEA DTVGCCSLRY EHVTLKPPNT VIFDFLGKDS IRFYQEVEVD KQVFKNLTIF 480
481 KRPPKQPGHQ LFDRLDPSIL NKYLQNYMPG LTAKVFRTYN ASKTMQDQLD LIPNKGSVAE 540
541 KILKYNAANR TVAILCNHQR TVTKGHAQTV EKANNRIQEL EWQKIRCKRA ILQLDKDLLK 600
601 KEPKYFEEID DLTKEDEATI HKRIIDREIE KYQRKFVREN DKRKFEKEEL LPESQLKEWL 660
661 EKVDEKKQEF EKELKTGEVE LKSSWNSVEK IKAQVEKLEQ RIQTSSIQLK DKEENSQVSL 720
721 GTSKINYIDP RLSVVFCKKY DVPIEKIFTK TLREKFKWAI ESVDENWRF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
NPL6 TOP1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
TOP1 TOP2 |
| View Details | Krogan NJ, et al. (2006) |
|
AIM26 APC9 BEM3 FMP27 IRA1 MDM36 MID1 NUP42 REV1 SEC20 SML1 TCO89 TOP1 YDR396W YGL024W ZAP1 ZIM17 |
| View Details | Gavin AC, et al. (2006) |
|
ADE5,7 REB1 TOP1 |
| View Details | Gavin AC, et al. (2002) |

|
ABF2 ADE5,7 CKA1 CKA2 CKB1 CKB2 CTI6 DEP1 DOT6 DPB2 DPB4 EAF3 ECM16 HHF1, HHF2 HTA2 IOC3 ISW1 ISW2 ITC1 KAP114 MED4 MOT1 NAP1 NHP10 NPL6 NRD1 PHO23 POL2 PUF6 RCO1 REB1 RET1 RFA1 RFX1 RNR2 RPA135 RPC34 RPC40 RPC82 RPD3 RPO31 RSC2 RSC3 RSC58 RSC8 RVB2 RXT2 RXT3 SAP30 SDS3 SFH1 SIN3 STH1 STO1 TAF1 TFC7 TOP1 TUP1 UME1 UME6 VPS1 YBR090C-A, NHP6B |
| View Details | Ho Y, et al. (2002) |
|
ACO1 AIM13 CLU1 RPC82 SPT16 TOP1 |
| View Details | Qiu et al. (2008) |

|
FOB1 TOP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
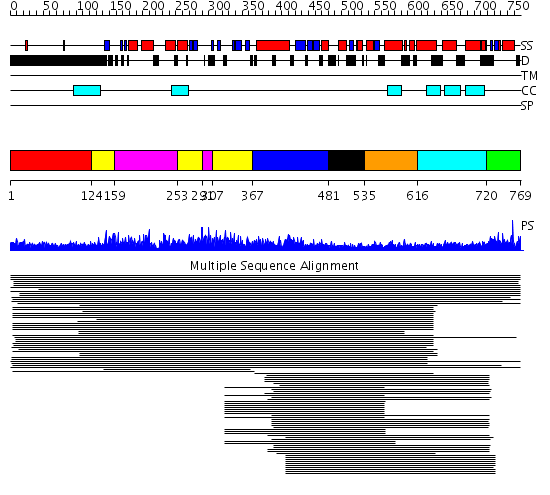
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..123] | 7.064997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [124..158] [253..290] [307..366] |
1352.0 | Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment | |
| 3 | View Details | [159..252] [291..306] |
1352.0 | Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment | |
| 4 | View Details | [367..480] | 57.9794 | Eukaryotic DNA topoisomerase I, catalytic core | |
| 5 | View Details | [481..534] | 3.221849 | Tropomyosin | |
| 6 | View Details | [535..615] | 3.221849 | Tropomyosin | |
| 7 | View Details | [616..719] | 3.221849 | Tropomyosin | |
| 8 | View Details | [720..769] | 12.79 | Eukaryotic DNA topoisomerase I, dispensable insert domain; Eukaryotic DNA topoisomerase I, catalytic core; Eukaryotic DNA topoisomerase I, N-terminal DNA-binding fragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)