













| General Information: |
|
| Name(s) found: |
VPS13 /
YLL040C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 3144 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA mitochondrion [IDA extrinsic to membrane [IDA |
| Biological Process: |
ascospore formation
[IMP protein retention in Golgi apparatus [IMP late endosome to vacuole transport [IGI vacuolar protein catabolic process [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLESLAANLL NRLLGSYVEN FDPNQLNVGI WSGDVKLKNL KLRKDCLDSL NLPIDVKSGI 60
61 LGDLVLTVPW SSLKNKPVKI IIEDCYLLCS PRSEDHENDE EMIKRAFRLK MRKVSEWELT 120
121 NQARILSTQS ENKTSSSSSE KNNAGFMQSL TTKIIDNLQV TIKNIHLRYE DMDGIFTTGP 180
181 SSVGLTLNEL SAVSTDSNWA PSFIDITQNI THKLLTLNSL CLYWNTDSPP LISDDDQDRS 240
241 LENFVRGFKD MIASKNSTAP KHQYILKPVS GLGKLSINKL GSTEEQPHID LQMFYDEFGL 300
301 ELDDTEYNDI LHVLSSIQLR QITKKFKKAR PSFAVSENPT EWFKYIAACV INEIHEKNKM 360
361 WTWESMKEKC EQRRLYTKLW VEKLKLKNLE APLRDPIQEA QLSELHKDLT YDEIILFRSV 420
421 AKRQYAQYKL GMTEDSPTPT ASSNIEPQTS NKSATKNNGS WLSSWWNGKP TEEVDEDLIM 480
481 TEEQRQELYD AIEFDENEDK GPVLQVPRER VELRVTSLLK KGSFTIRKKK QNLNLGSIIF 540
541 ENCKVDFAQR PDSFLSSFQL NKFSLEDGSP NALYKHIISV RNSSKDQSSI DNHATGEEEE 600
601 EDEPLLRASF ELNPLDGLAD SNLNIKLLGM TVFYHVHFIT EVHKFFKASN QHMETIGNIV 660
661 NAAEATVEGW TTQTRMGIES LLEDHKTVNV SLDLQAPLII LPLDPHDWDT PCAIIDAGHM 720
721 SILSDLVPKE KIKEIKELSP EEYDKIDGNE INRLMFDRFQ ILSQDTQIFV GPDIQSTIGK 780
781 INTASSTNDF RILDKMKLEL TVDLSILPKA YKLPTIRVFG HLPRLSLSIN DIQYKTIMNL 840
841 IANSIPSMID DEENNGDYVN YSSGSEKEMK KQIQLQLKNT LKALENMQPL QIEQKFLELH 900
901 FDIDQAKIAF FQCIKNDSRN SEKLVDILCQ RLNFNFDKRA KEMNLDLRVH SLDVEDYIEL 960
961 TDNKEFKNLI SSGVEKVTRS QKDLFTLKYK RVQRIVPHND TLIELFDQDI VMHMSELQLV1020
1021 LTPRSVLTLM NYAMLTFTDP NAPEMPADVL RHNKEDRDDA PQKINMKIKM EAVNVIFNDD1080
1081 SIKLATLVLS AGEFTMVLLP ERYNINLKLG GLELTDETNE SFSRDSVFRK IIQMKGQELV1140
1141 ELSYESFDPA TNTKDYDSFL KYSTGSMHVN FIESAVNRMV NFFAKFQKSK VSFDRARLAA1200
1201 YNQAPSIDAV NNMKMDIVIK APIIQFPKLV GTQENNYDTM RFYLGEFFIE NKFSVIDESH1260
1261 KINHIKLGVR EGQLSSNLNF DGSSQQLYLV ENIGLLFNID RDPLPQDDTP ELKVTSNFES1320
1321 FALDLTENQL TYLLEISNKV SSAFNITDEN SGESGGKGEI KSPSPDPASL SSESERTATP1380
1381 QSLQGSNKSN IKNPEQKYLD FSFKAPKIAL TLYNKTKGVT SLNDCGLTRI MFQDIGCSLG1440
1441 LKNDGTVDGQ AHVAAFRIED VRNIKDNKHT ELIPKSKNKE YQFVANISRK NLEVGRLLNI1500
1501 SMTMDSPKMI LAMDYLVSLK EFFDAIMSKS HENNLYYPEN TNQKPENKAI VESVQEGGDV1560
1561 TKIQYSVNII ETALILLADP CDMNSEAISF KIGQFLVTDQ NIMTVAANNV GIFLFKMNSS1620
1621 EEKLRLLDDF SSSLTIDKRN STPQTLMTNI QLSVQPLLMR ISLRDIRLAM LIFKRVTTLL1680
1681 NKMTEKEDNG EEEESTDKIQ FSHEFERKLA VLDPSILGER SRASQSSDSE SIEVPTAILK1740
1741 NETFNADLGG LRFILIGDVH EMPILDMNVN EITASAKDWS TDFEALASLE TYVNIFNYSR1800
1801 SSWEPLLEMI PITFHLSKGH SEMDPAFSFD ILTQRIAEIT LSARSIAMLS HIPASLTEEL1860
1861 PLASRVSQKP YQLVNDTELD FDVWIQDKTT EDNKNEVVLL KANTSLPWEF EDWRSIREKL1920
1921 DIDKSKNILG VCVSGQNYKT IMNIDATTEG ENLHVLSPPR NNVHNRIVCE ARCDENNVKI1980
1981 ITFRSTLVIE NTTSTEIELL VDSKDPNKPS LKYAIKPHQS KSVPVEYAYD SDIRIRPASE2040
2041 DIYDWSQQTL SWKSLLSNQM SIFCSSKEDS NQRFHFEIGA KYDEREPLAK IFPHMKIVVS2100
2101 ASMTIENLLP ADINFSIFDK REEKRTDFLK TGESMEVHHI SLDSFLLMSV QPLQDEASAS2160
2161 KPSIVNTPHK SPLNPEDSLS LTLSGGQNLL LKLDYKNIDG TRSKVIRIYS PYIIMNSTDR2220
2221 ELYIQSSLLN IAQSKILLEN EKRYTIPKMF SFDKEDDKSN RARIRFKESE WSSKLSFDAI2280
2281 GQSFDASVRI KNKEQESNLG INISEGKGKY LLSKVIEIAP RYIISNTLDI PIEVCETGSM2340
2341 DVQQIESNIT KPLYRMRNIV DKQLVLKFLG GDSNWSQPFF IKNVGVTYLK VLKNSRHKLL2400
2401 KIEILLDKAT IFIRIKDGGD RWPFSIRNFS DHDFIFYQRD PRKVSDPYKD DQSNESSSRS2460
2461 FKPIFYRIPS KSIMPYAWDF PTAKEKYLVL ESGTRTREVR LAEIGELPPL RLDKRSKDKP2520
2521 APIVGLHVVA DDDMQALVIV NYKANVGLYK LKTASATTTS SVSVNSSVTD GFVQKDEDEK2580
2581 VNTQIVVSFK GVGISLINGR LQELLYINMR GIELRYNESK AYQTFSWKMK WMQIDNQLFS2640
2641 GNYSNILYPT EIPYTEKEIE NHPVISGSIS KVNDSLQAVP YFKHVTLLIQ EFSIQLDEDM2700
2701 LYAMMDFIKF PGSPWIMDSR DYKYDEEIQL PDVSELKTAG DIYFEIFHIQ PTVLHLSFIR2760
2761 SDEISPGLAE ETEESFSSSL YYVHMFAMTL GNINEAPVKV NSLFMDNVRV PLPILMDHIE2820
2821 RHYTTQFVYQ IHKILGSADC FGNPVGLFNT ISSGVWDLFY EPYQGYMMND RPQEIGIHLA2880
2881 KGGLSFAKKT VFGLSDSMSK FTGSMAKGLS VTQDLEFQRV RRLQQRINKN NRNALANSAQ2940
2941 SFASTLGSGL SGIALDPYKA MQKEGAAGFL KGLGKGIVGL PTKTAIGFLD LTSNLSQGVK3000
3001 STTTVLDMQK GCRVRLPRYV DHDQIIKPYD LREAQGQYWL KTVNGGVFMN DEYLSHVILP3060
3061 GKELAVIVSM QHIAEVQMAT QELMWSTGYP SIQGITLERS GLQIKLKSQS EYFIPISDPE3120
3121 ERRSLYRNIA IAVREYNKYC EAIL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
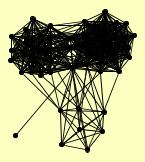
| View Details | Krogan NJ, et al. (2006) |
|
MUM2 VPS13 |
| View Details | Gavin AC, et al. (2002) |

|
CTF18 CTF8 DPB2 ELG1 OLA1 POL2 RFC1 RFC2 RFC3 RFC4 RFC5 TOP2 VPS13 YMR181C |
| View Details | Ho Y, et al. (2002) |
|
ADR1 BBC1 BCY1 CDC39 CDC53 CMD1 CMK2 CMP2 COF1 CPR1 CRM1 DUR1,2 ECM29 ECM33 EDE1 FAA4 FET4 GAL3 GCN1 HCH1 HRT1 HYP2 ILS1 IPP1 MDN1 MKT1 MMS1 MYO2 MYO4 MYO5 PFK1 PGM2 PMA2 PST2 RIM15 RPA190 RPN1 SHE3 SOD1 TCP1 TPK1 TPK2 TPK3 TPS1 UBA1 UBI4 VAS1 VPS13 YAR009C YGP1 YLR035C-A YNK1 YPT35 |
The following runs contain data for this protein:
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
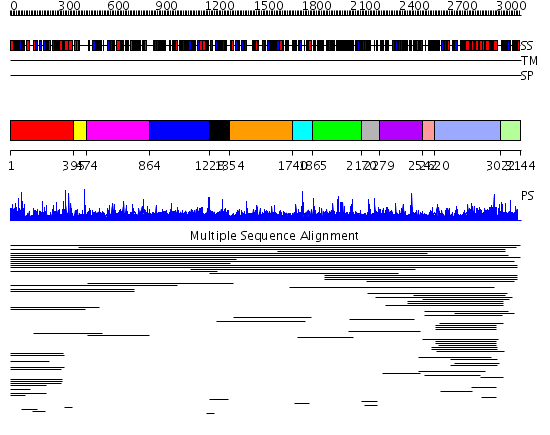
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..394] | 5.024958 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [395..473] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [474..863] | 1.016912 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [864..1227] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1228..1353] | 1.015977 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1354..1739] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1740..1864] | 1.015982 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1865..2169] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [2170..2278] | 2.019983 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [2279..2541] | 1.020955 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [2542..2619] | N/A | Confident ab initio structure predictions are available. | |
| 12 | View Details | [2620..3021] | 4.033932 | View MSA. No confident structure predictions are available. | |
| 13 | View Details | [3022..3144] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)