













| General Information: |
|
| Name(s) found: |
PRP5 /
YBR237W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 849 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spliceosomal complex
[TAS |
| Biological Process: |
RNA splicing
[IEA]
nuclear mRNA branch site recognition [TAS] mRNA processing [IEA] |
| Molecular Function: |
ATP-dependent RNA helicase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METIDSKQNI NRESLLEERR KKLAKWKQKK AQFDAQKEHQ TSRNDIVTNS LEGKQTTEKF 60
61 TERQERVKEE LRKRKNEFRK SDEPVSVKPS KKKSKRSKVK KKISFDFSDD DDSEIGVSFR 120
121 SKEHIQKAPE HDNEKDPLDE FMTSLKEEKM SNSKGMYDRG DILDVEDQLF ELGGTDDEDV 180
181 EDNTDNSNIA KIAKLKAKKR VKQIYYSPEE LEPFQKNFYI ESETVSSMSE MEVEELRLSL 240
241 DNIKIKGTGC PKPVTKWSQL GLSTDTMVLI TEKLHFGSLT PIQSQALPAI MSGRDVIGIS 300
301 KTGSGKTISY LLPLLRQVKA QRPLSKHETG PMGLILAPTR ELALQIHEEV TKFTEADTSI 360
361 RSVCCTGGSE MKKQITDLKR GTEIVVATPG RFIDILTLND GKLLSTKRIT FVVMDEADRL 420
421 FDLGFEPQIT QIMKTVRPDK QCVLFSATFP NKLRSFAVRV LHSPISITIN SKGMVNENVK 480
481 QKFRICHSED EKFDNLVQLI HERSEFFDEV QSENDGQSSD VEEVDAKAII FVSSQNICDF 540
541 ISKKLLNAGI VTCAIHAGKP YQERLMNLEK FKREKNSILL CTEVLSRGLN VPEVSLVIIY 600
601 NAVKTFAQYV HTTGRTARGS RSGTAITLLL HDELSGAYIL SKAMRDEEIK ALDPLQAKEL 660
661 QEMSAKFESG MKKGKFRLSK GFGGKGLENI KSKREEAQNK DLELKKNDKR SDDLEKKISN 720
721 PREGHDSVSE SSALIPRLNY ELFKESTDGS IIFYAKVYIN DLPQIVRWEA TKNTTLLFIK 780
781 HETGCSITNK GKFYPEGKEP KNENDEPKLY LLIEGQDEKD IQLSIELLEQ KVKEGVVKAA 840
841 SLSLKSTKY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
AAR2 BRR1 BRR2 CBC2 CDC40 CWC15 CWC2 CWC22 CWC23 DHR2 LSM4 MAK5 MUD2 PRP16 PRP18 PRP2 PRP22 PRP24 PRP28 PRP31 PRP38 PRP39 PRP4 PRP40 PRP43 PRP5 PRP6 PRP8 SLU7 SMD1 SMD3 SME1 SNP1 SPP2 SPP381 STO1 YHC1 |
| View Details | Krogan NJ, et al. (2006) |

|
ADE2 BSC2 CWH41 DBF4 DIG1 ECM29 HAL5 LAA1 MRC1 OPI9 PEX32 PRP5 PTH2 RGA2 RMA1 RPN3 SIR1 STE12 THR1 TLG2 VPS45 YDL199C YEL057C YPL035C |
| View Details | Qiu et al. (2008) |

|
AAR2 BRR2 BUD31 CBC2 CDC40 CEF1 CLF1 CUS1 CUS2 CWC15 CWC2 CWC21 CWC22 CWC23 CWC25 DIB1 ECM2 HSH155 IST3 ISY1 LEA1 LSM2 LSM3 LSM4 LSM5 LSM7 LSM8 LUC7 MAK5 MSL1 MUD1 MUD2 NAM8 NTC20 PRP11 PRP16 PRP18 PRP19 PRP2 PRP21 PRP22 PRP24 PRP28 PRP3 PRP31 PRP38 PRP39 PRP4 PRP40 PRP42 PRP43 PRP45 PRP46 PRP5 PRP6 PRP8 PRP9 RSE1 SLU7 SMB1 SMD1 SMD2 SMD3 SME1 SMX2 SMX3 SNP1 SNT309 SNU114 SNU23 SNU56 SNU66 SNU71 SPP2 SPP381 SPP382 STO1 SUB2 SYF1 SYF2 YHC1 YJU2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
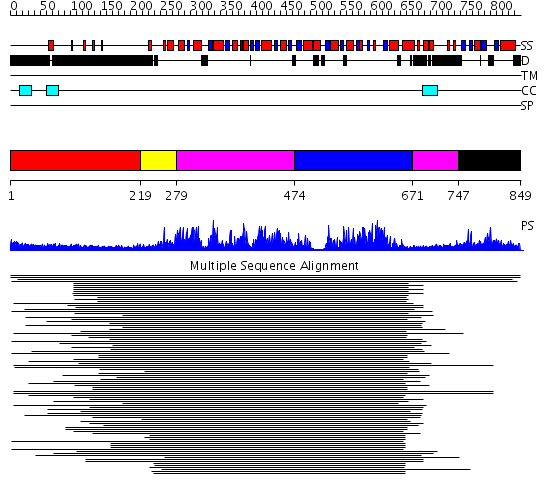
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 8.274997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [219..278] | 461.0 | Initiation factor 4a | |
| 3 | View Details | [279..473] [671..746] |
13.30103 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [474..670] | 13.30103 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [747..849] | 8.62 | RNA splicing factor 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)