GO TERM SUMMARY
|
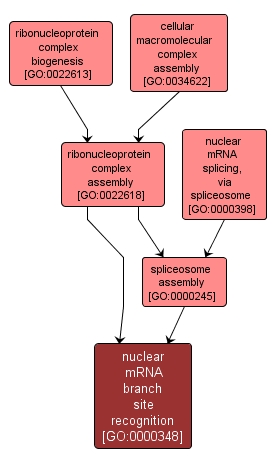
| Name: |
nuclear mRNA branch site recognition |
| Acc: |
GO:0000348 |
| Aspect: |
Biological Process |
| Desc: |
Recognition of the pre-mRNA branch site sequence by components of the assembling spliceosome. |
Synonyms:
- spliceosomal B complex biosynthesis
- GO:0000371
- GO:0000370
- U12-type nuclear mRNA branch site recognition
- U2-type nuclear mRNA branch site recognition
- spliceosomal A complex formation
- spliceosomal B complex formation
- spliceosomal A complex biosynthesis
|