













| General Information: |
|
| Name(s) found: |
TUB3 /
YML124C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 445 amino acids |
Gene Ontology: |
|
| Cellular Component: |
tubulin complex
[TAS]
kinetochore microtubule [TAS spindle pole body [IDA polar microtubule [TAS nuclear microtubule [TAS cytoplasmic microtubule [TAS |
| Biological Process: |
homologous chromosome segregation
[TAS mitotic sister chromatid segregation [TAS nuclear migration along microtubule [TAS nuclear migration during conjugation with cellular fusion [TAS |
| Molecular Function: |
structural constituent of cytoskeleton
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MREVISINVG QAGCQIGNAC WELYSLEHGI KEDGHLEDGL SKPKGGEEGF STFFHETGYG 60
61 KFVPRAIYVD LEPNVIDEVR TGRFKELFHP EQLINGKEDA ANNYARGHYT VGREIVDEVE 120
121 ERIRKMADQC DGLQGFLFTH SLGGGTGSGL GSLLLENLSY EYGKKSKLEF AVYPAPQLST 180
181 SVVEPYNTVL TTHTTLEHAD CTFMVDNEAI YDICKRNLGI SRPSFSNLNG LIAQVISSVT 240
241 ASLRFDGSLN VDLNEFQTNL VPYPRIHFPL VSYAPILSKK RATHESNSVS EITNACFEPG 300
301 NQMVKCDPTK GKYMANCLLY RGDVVTRDVQ RAVEQVKNKK TVQMVDWCPT GFKIGICYEP 360
361 PSVIPSSELA NVDRAVCMLS NTTAIADAWK RIDQKFDLMY AKRAFVHWYV GEGMEEGEFT 420
421 EAREDLAALE RDYIEVGADS YAEEF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | Boi 2 gst control | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
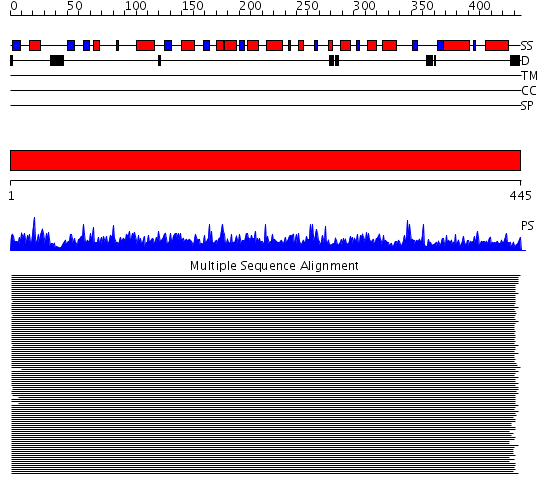
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..445] | 11000.0 | Tubulin:stathmin-like domain complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)