













| General Information: |
|
| Name(s) found: |
PRP46 /
YPL151C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 451 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spliceosomal complex
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGNDHKVEN LGDVDKFYSR IRWNNQFSYM ATLPPHLQSE MEGQKSLLMR YDTYRKESSS 60
61 FSGEGKKVTL QHVPTDFSEA SQAVISKKDH DTHASAFVNK IFQPEVAEEL IVNRYEKLLS 120
121 QRPEWHAPWK LSRVINGHLG WVRCVAIDPV DNEWFITGSN DTTMKVWDLA TGKLKTTLAG 180
181 HVMTVRDVAV SDRHPYLFSV SEDKTVKCWD LEKNQIIRDY YGHLSGVRTV SIHPTLDLIA 240
241 TAGRDSVIKL WDMRTRIPVI TLVGHKGPIN QVQCTPVDPQ VVSSSTDATV RLWDVVAGKT 300
301 MKVLTHHKRS VRATALHPKE FSVASACTDD IRSWGLAEGS LLTNFESEKT GIINTLSINQ 360
361 DDVLFAGGDN GVLSFYDYKS GHKYQSLATR EMVGSLEGER SVLCSTFDKT GLRLITGEAD 420
421 KSIKIWKQDE TATKESEPGL AWNPNLSAKR F |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
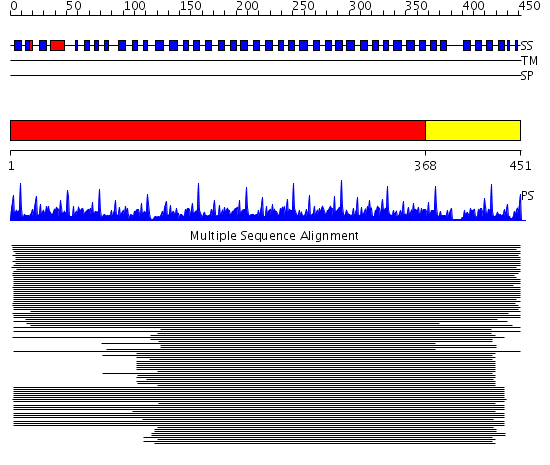
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..367] | 214.0 | Cdc4 F-box and linker domains; Cdc4 propeller domain | |
| 2 | View Details | [368..451] | 218.54902 | Tup1, C-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)