













| General Information: |
|
| Name(s) found: |
PDB1 /
YBR221C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 366 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial pyruvate dehydrogenase complex
[TAS mitochondrion [IDA mitochondrial nucleoid [IDA |
| Biological Process: |
pyruvate metabolic process
[NAS |
| Molecular Function: |
pyruvate dehydrogenase (acetyl-transferring) activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSRLPTSLA RNVARRAPTS FVRPSAAAAA LRFSSTKTMT VREALNSAMA EELDRDDDVF 60
61 LIGEEVAQYN GAYKVSKGLL DRFGERRVVD TPITEYGFTG LAVGAALKGL KPIVEFMSFN 120
121 FSMQAIDHVV NSAAKTHYMS GGTQKCQMVF RGPNGAAVGV GAQHSQDFSP WYGSIPGLKV 180
181 LVPYSAEDAR GLLKAAIRDP NPVVFLENEL LYGESFEISE EALSPEFTLP YKAKIEREGT 240
241 DISIVTYTRN VQFSLEAAEI LQKKYGVSAE VINLRSIRPL DTEAIIKTVK KTNHLITVES 300
301 TFPSFGVGAE IVAQVMESEA FDYLDAPIQR VTGADVPTPY AKELEDFAFP DTPTIVKAVK 360
361 EVLSIE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
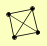
|
LAT1 LPD1 PDA1 PDB1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
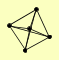
|
LAT1 LPD1 PDA1 PDB1 PDX1 |
| View Details | Krogan NJ, et al. (2006) |
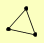
|
ADY3 AIM23 PDB1 |
| View Details | Gavin AC, et al. (2006) |
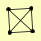
|
LAT1 PDA1 PDB1 PDX1 |
| View Details | Gavin AC, et al. (2002) |
|
APM1 AVO1 BEM2 BLM10 CEG1 CET1 CLU1 CTR9 CYM1 DAM1 DBP9 DOA4 DUR1,2 ECM29 EST3 GFA1 HHF1, HHF2 HSP42 INO4 KAP95 LAT1 LPD1 LSM12 LYS12 MDS3 NIP1 NOP1 NTE1 NUD1 PDA1 PDB1 PDX1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE8 PRE9 PRP43 PRP46 PSE1 PUP3 RGR1 RPG1 RPN11 RPT3 RPT5 RRP12 SCL1 SLX9 SPA2 SPT16 SRP1 SSA3 SSA4 TCM62 TOP2 ULP1 VPS1 YFL006W YMR310C YRA1 |
| View Details | Qiu et al. (2008) |
|
KGD1 LPD1 PDA1 PDB1 PDX1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | De Craene, J., et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
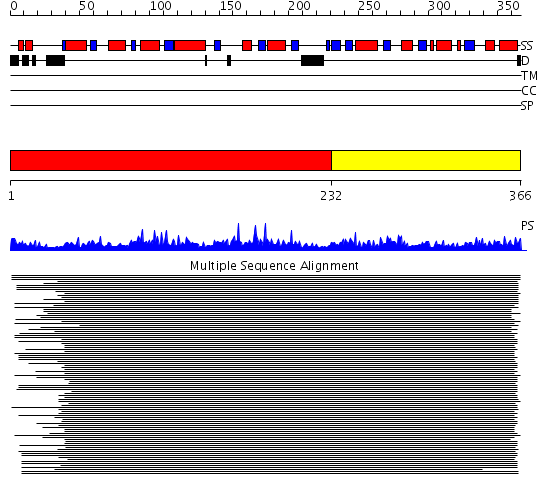
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 626.855515 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain | |
| 2 | View Details | [232..366] | 626.855515 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain |
Functions predicted (by domain):
| Source: Reynolds et al. 2008. Manuscript submitted |

|