













| General Information: |
|
| Name(s) found: |
NTE1 /
YML059C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1679 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA |
| Biological Process: |
sinus venosus formation
[IDA phosphatidylcholine metabolic process [IMP |
| Molecular Function: |
lysophospholipase activity
[IDA hydrolase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRSMNCTTNN TNNTGQNTKN SLGSSFNSSN YTSYRFQTCL TDQIISEAQT WSLSSLFNFS 60
61 WVVSYFVMGA SRMIFRYGWY LATLSLLRIP KWIFFKLHHV QFTLSFWLIL FALAVIVFVT 120
121 YTIMKERILS QYKRLTPEFL PLENTGKSGS SANINAASTQ SANAPPAIGS STTGASSIID 180
181 SKKHSLKDGN ENETFLSSYL DQFLSAIKIF GYLEKPVFHD LTKNMKTQKM DEGEILLLDS 240
241 TIGFAIVVEG TLQLYHEVDH SDKDHGDETD HSDTDGLDDQ DRDEEDEEED DDIDNYDTKS 300
301 CSSNLIDEED ESVGYIHLKN GLGNFQLLNT VKPGNPLTSL VSILNLFTHS MSSYGNSNFP 360
361 SELSSPIDTT VSVNNMFCSS EQNFSNTDSM TNSTNSFPTF PSSMPKLVAR AATDCTIGII 420
421 PPQSFAKLTA KYPRSASHII QMVLTKLYHV TFQTAHDYLG LTKEIMDIEV LLNKSIVYEL 480
481 PYYLKEAVIR KFKTVDKSSG SADLEPKPKN SNASSKLKKP PKAKPSDGII QSLKIANANA 540
541 NTSSNSLSLK PEFTHHPSSR HVVLGSRDQF NPGDLLSNVP LSRTMDILSP NPIHNNNRNK 600
601 SNGINTSTSN QHKRSSRSSS NNASVHSKKF SSLSPELRNA QLSTSPLSLD NTSVHDHIHP 660
661 SPVHLKGRVS PRPNLLPTTS FSAAQEETED SALRMALVEA MLTYLGVNKS NMSVSSSSIA 720
721 NMSSLNSPQL NEMYSRRPSN ASFLMSPHCT PSDISVASSF ASPQTQPTML RILPKEYTIS 780
781 NKRHNKSKSQ DKKKPRAYKE ELTPNLDFED VKKDFAQGIQ LKFFKKGTTI VEQNARGKGL 840
841 FYIISGKVNV TTNSSSSVVS SMSKPEQVSA QSSHKGENPH HTQHLLYSVG SGGIVGYLSS 900
901 LIGYKSFVNI VAKSDVYVGF LSSATLERLF DKYFLIYLRI SDSLTKLLSS RLLKLDHALE 960
961 WVHLRASETL FSQGDSANGI YVVLNGRLRQ LQQQSLSNSN TSSEEVETQN IILGELAQGE1020
1021 SFGEVEVLTA MNRYSTIVAV RDSELARIPR TLFELLALEH PSIMIRVSRL VAKKIVGDRT1080
1081 VPALTGDPLS IKENDFTSLI PPTKASYSSS LSHKPQNITS GTITFRTITI LPITSGLPVE1140
1141 AFAMKLVQAF KQVGRTTIGL NQRTTLTHLG RHAFDRLSKL KQSGYFAELE EMYQTVVYIS1200
1201 DTPVKSNWTR TCIAQGDCIL LLADARSPSA EIGEYEKLLL NSKTTARTEL ILLHPERYVE1260
1261 PGLTHKWLRY RPWVHSHHHI QFSLTGTTLM NEGKMHVLNN GALALMDKLI QTEFSRKTQQ1320
1321 NISKLLPDSI KNTVENFSSR FMKSKRQYYT PVHRHKNDFL RLARILSGQA IGLVLGGGGA1380
1381 RGISHLGVIQ AIEEQGIPVD VIGGTSIGSF VGGLYAKDYD LVPIYGRVKK FAGRISSIWR1440
1441 MLTDLTWPVT SYTTGHEFNR GIWKTFGDTR IEDFWIQYYC NSTNITDSVQ EIHSFGYAWR1500
1501 YIRASMSLAG LLPPLEENGS MLLDGGYVDN LPVTEMRARG CQTIFAVDVG SADDRTPMEY1560
1561 GDSLNGFWII FNRWNPFSSH PNIPNMAEIQ VRLGYVASVN ALEKAKNTPG VVYVRPPIEE1620
1621 YATLDFSKFE EIYHVGVDYG RIFLQGLIDD DKMPYIPGSQ ETTLNSQVPE FLLHRRNSI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
MSS1 NOP7 NTE1 PDS5 SOP4 STP3 YGR067C YTM1 |
| View Details | Gavin AC, et al. (2002) |
|
NTE1 PDB1 PSE1 |
| View Details | Qiu et al. (2008) |

|
BST1 NTE1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
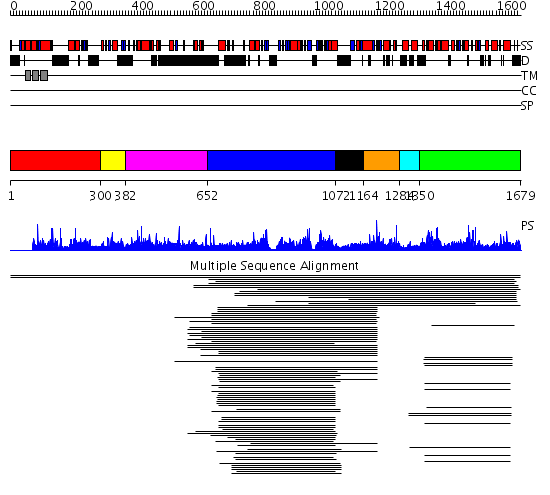
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [300..381] | 1.002995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [382..651] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [652..1071] | 138.616428 | No description for 1apk__ was found. | |
| 5 | View Details | [1072..1163] | 47.0 | No description for 2gapA_ was found. | |
| 6 | View Details | [1164..1283] | 1.170988 | View MSA. Confident ab initio structure predictions are available. | |
| 7 | View Details | [1284..1349] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1350..1679] | 52.142993 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1552..1679] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|