













| General Information: |
|
| Name(s) found: |
MSS1 /
YMR023C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 526 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
tRNA modification
[IMP translation [IMP |
| Molecular Function: |
GTP binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSASFLQSR LISRSFLVRR SLKRYSGLAK PYTFQQPTIY ALSTPANQTS AIAIIRISGT 60
61 HAKYIYNRLV DSSTVPPIRK AILRNIYSPS SCSVKPHDQK ESKILLDTSL LLYFQAPYSF 120
121 TGEDVLELHV HGGKAVVNSI LKAIGSLHDR SSGKDIRFAL PGDFSRRAFQ NGKFDLTQLE 180
181 GIKDLIDSET ESQRRSALSS FNGDNKILFE NWRETIIENM AQLTAIIDFA DDNSQEIQNT 240
241 DEIFHNVEKN IICLRDQIVT FMQKVEKSTI LQNGIKLVLL GAPNVGKSSL VNSLTNDDIS 300
301 IVSDIPGTTR DSIDAMINVN GYKVIICDTA GIREKSSDKI EMLGIDRAKK KSVQSDLCLF 360
361 IVDPTDLSKL LPEDILAHLS SKTFGNKRII IVVNKSDLVS DDEMTKVLNK LQTRLGSKYP 420
421 ILSVSCKTKE GIESLISTLT SNFESLSQSS ADASPVIVSK RVSEILKNDV LYGLEEFFKS 480
481 KDFHNDIVLA TENLRYASDG IAKITGQAIG IEEILDSVFS KFCIGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
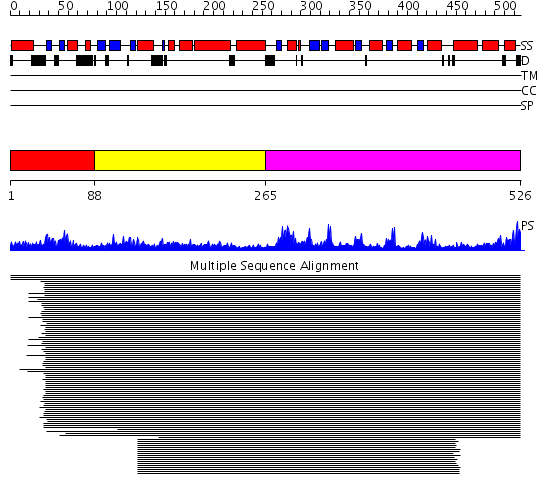
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 1.105991 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [88..264] | 2.660996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [265..526] | 125.9794 | GTPase Era, N-terminal domain; GTPase Era C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)